Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
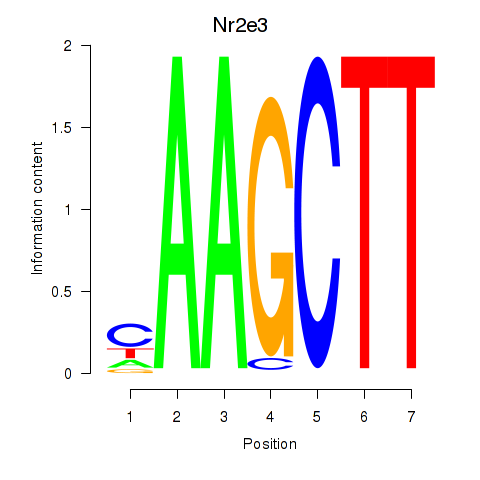
Results for Nr2e3
Z-value: 1.05
Transcription factors associated with Nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e3
|
ENSRNOG00000050690 | nuclear receptor subfamily 2, group E, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e3 | rn6_v1_chr8_-_64800467_64800467 | 0.15 | 8.6e-03 | Click! |
Activity profile of Nr2e3 motif
Sorted Z-values of Nr2e3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_118108864 | 51.99 |
ENSRNOT00000006184
|
Mb
|
myoglobin |
| chr18_-_28017925 | 51.89 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chrX_-_142164220 | 44.64 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr15_-_14737704 | 44.56 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr4_+_175729726 | 41.73 |
ENSRNOT00000013230
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr8_-_80631873 | 40.80 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr4_-_727691 | 31.24 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr17_-_61332391 | 31.09 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr9_-_85243001 | 30.61 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr10_-_27366665 | 29.27 |
ENSRNOT00000004725
|
Gabra1
|
gamma-aminobutyric acid type A receptor alpha1 subunit |
| chr3_+_137618898 | 28.78 |
ENSRNOT00000007249
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr3_+_129599353 | 26.21 |
ENSRNOT00000008734
|
Snap25
|
synaptosomal-associated protein 25 |
| chr15_-_6587367 | 25.99 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chr11_-_71750014 | 25.76 |
ENSRNOT00000072227
|
LOC100911042
|
uncharacterized LOC100911042 |
| chr2_+_3750094 | 25.63 |
ENSRNOT00000064743
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr14_-_112946875 | 25.26 |
ENSRNOT00000081981
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr4_-_64330996 | 24.56 |
ENSRNOT00000016088
|
Ptn
|
pleiotrophin |
| chr8_+_53411316 | 23.35 |
ENSRNOT00000011107
ENSRNOT00000086957 |
Tmprss5
|
transmembrane protease, serine 5 |
| chrX_+_92596378 | 22.20 |
ENSRNOT00000045001
|
Pcdh11x
|
protocadherin 11 X-linked |
| chr4_-_29778039 | 20.63 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr1_+_25174456 | 20.44 |
ENSRNOT00000092830
|
Clvs2
|
clavesin 2 |
| chr6_+_110410141 | 19.93 |
ENSRNOT00000013867
|
Esrrb
|
estrogen-related receptor beta |
| chr8_+_70603249 | 19.37 |
ENSRNOT00000067016
ENSRNOT00000072486 |
Igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chrX_+_20423401 | 19.01 |
ENSRNOT00000093162
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr4_+_130332076 | 18.65 |
ENSRNOT00000083127
|
Mitf
|
melanogenesis associated transcription factor |
| chr3_+_58164931 | 18.48 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr10_+_53740841 | 17.50 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr10_-_18579632 | 16.94 |
ENSRNOT00000040737
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr8_+_107882219 | 16.88 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr15_+_18399733 | 16.59 |
ENSRNOT00000061158
|
Fam107a
|
family with sequence similarity 107, member A |
| chr14_-_17582823 | 16.18 |
ENSRNOT00000087686
|
LOC100911949
|
uncharacterized LOC100911949 |
| chr15_+_18399515 | 14.79 |
ENSRNOT00000071273
|
Fam107a
|
family with sequence similarity 107, member A |
| chr6_-_67084234 | 14.46 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr8_+_114897011 | 14.24 |
ENSRNOT00000074683
|
Twf2
|
twinfilin actin-binding protein 2 |
| chr16_+_39909270 | 13.60 |
ENSRNOT00000081994
|
Wdr17
|
WD repeat domain 17 |
| chr8_+_122076759 | 13.20 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr3_-_79728879 | 12.70 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr4_+_24222500 | 12.61 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr3_-_48535909 | 12.52 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr13_+_60435946 | 12.52 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr7_+_64672722 | 12.35 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr7_+_123102493 | 11.82 |
ENSRNOT00000038612
|
Aco2
|
aconitase 2 |
| chr15_-_4057104 | 11.03 |
ENSRNOT00000084350
ENSRNOT00000012270 |
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr1_-_178195948 | 10.86 |
ENSRNOT00000081767
|
Btbd10
|
BTB domain containing 10 |
| chr17_-_55300998 | 10.42 |
ENSRNOT00000064331
|
Svil
|
supervillin |
| chr6_-_76552559 | 10.37 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr2_-_22385855 | 10.19 |
ENSRNOT00000083531
|
Thbs4
|
thrombospondin 4 |
| chr10_-_40953467 | 10.12 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chrX_-_112473822 | 9.74 |
ENSRNOT00000079180
|
Col4a6
|
collagen type IV alpha 6 chain |
| chr5_+_122019301 | 9.50 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr2_+_247248407 | 9.01 |
ENSRNOT00000082287
|
Unc5c
|
unc-5 netrin receptor C |
| chrX_+_15669191 | 8.82 |
ENSRNOT00000013553
|
Magix
|
MAGI family member, X-linked |
| chrX_-_104932508 | 8.80 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr4_+_163293724 | 8.70 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr17_-_15467320 | 8.44 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr11_-_81379640 | 8.43 |
ENSRNOT00000002484
|
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr6_-_76608864 | 8.32 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr16_-_82439441 | 8.30 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr11_+_60907015 | 7.96 |
ENSRNOT00000002797
|
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr2_-_250232295 | 7.84 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr14_+_32257805 | 7.79 |
ENSRNOT00000039626
|
AABR07014804.1
|
|
| chr14_-_21127868 | 7.52 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr2_+_184230459 | 7.46 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr2_-_177950517 | 7.12 |
ENSRNOT00000012792
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr5_-_169017295 | 6.96 |
ENSRNOT00000067481
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr19_+_37229120 | 6.59 |
ENSRNOT00000076335
|
Hsf4
|
heat shock transcription factor 4 |
| chr7_-_144319804 | 6.44 |
ENSRNOT00000070855
|
LOC100912282
|
calcium-binding and coiled-coil domain-containing protein 1-like |
| chr10_+_81913689 | 6.44 |
ENSRNOT00000003780
|
Tob1
|
transducer of ErbB-2.1 |
| chr3_-_134696654 | 6.43 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr1_+_98627372 | 6.01 |
ENSRNOT00000030370
|
Dzf17
|
zinc finger protein 17 |
| chr3_+_2158995 | 5.43 |
ENSRNOT00000010270
|
Zmynd19
|
zinc finger, MYND-type containing 19 |
| chr2_-_211831551 | 5.41 |
ENSRNOT00000039225
ENSRNOT00000090561 |
Fam102b
|
family with sequence similarity 102, member B |
| chr5_-_128186532 | 5.36 |
ENSRNOT00000012635
|
Prpf38a
|
pre-mRNA processing factor 38A |
| chrX_+_1787266 | 4.72 |
ENSRNOT00000011183
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr3_+_56802714 | 4.02 |
ENSRNOT00000077551
|
Erich2
|
glutamate-rich 2 |
| chrX_-_25628272 | 3.71 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr20_+_4363508 | 3.66 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chr2_+_58462588 | 3.64 |
ENSRNOT00000083799
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr1_-_78521999 | 3.42 |
ENSRNOT00000021223
|
Arhgap35
|
Rho GTPase activating protein 35 |
| chr8_-_64154396 | 3.35 |
ENSRNOT00000031262
|
Bbs4
|
Bardet-Biedl syndrome 4 |
| chr1_-_220974597 | 3.21 |
ENSRNOT00000088799
|
Kat5
|
lysine acetyltransferase 5 |
| chr14_+_63095720 | 3.16 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr18_+_30808404 | 3.08 |
ENSRNOT00000060475
|
Pcdhga1
|
protocadherin gamma subfamily A, 1 |
| chr4_-_28224858 | 2.82 |
ENSRNOT00000091075
|
AABR07059632.3
|
|
| chr19_-_17494516 | 2.44 |
ENSRNOT00000078786
ENSRNOT00000076138 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr18_+_28370085 | 2.43 |
ENSRNOT00000083993
|
Matr3
|
matrin 3 |
| chr3_-_157099306 | 2.42 |
ENSRNOT00000022861
|
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr1_+_237574788 | 2.33 |
ENSRNOT00000042738
|
AABR07006504.1
|
|
| chr9_+_2202511 | 2.25 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr1_-_126227469 | 2.04 |
ENSRNOT00000087332
|
Tjp1
|
tight junction protein 1 |
| chr1_+_170147300 | 1.47 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr18_+_29506128 | 1.47 |
ENSRNOT00000003878
|
Slc35a4
|
solute carrier family 35, member A4 |
| chr6_+_10533151 | 1.03 |
ENSRNOT00000020822
|
Rhoq
|
ras homolog family member Q |
| chr4_+_1371401 | 0.73 |
ENSRNOT00000071676
|
LOC100910899
|
olfactory receptor 143-like |
| chr10_-_95262024 | 0.70 |
ENSRNOT00000066487
|
Bptf
|
bromodomain PHD finger transcription factor |
| chr16_+_81665540 | 0.69 |
ENSRNOT00000026546
ENSRNOT00000092644 ENSRNOT00000073240 |
Grtp1
|
growth hormone regulated TBC protein 1 |
| chr3_+_79728796 | 0.64 |
ENSRNOT00000068124
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr3_-_175654707 | 0.60 |
ENSRNOT00000080191
|
Rbbp8nl
|
RBBP8 N-terminal like |
| chr20_-_32258286 | 0.59 |
ENSRNOT00000082114
|
Ddx50
|
DExD-box helicase 50 |
| chr5_+_107233230 | 0.35 |
ENSRNOT00000029976
|
Ifne
|
interferon, epsilon |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.3 | 52.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 10.4 | 31.2 | GO:0060769 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 8.7 | 26.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 6.5 | 51.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 6.4 | 44.6 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 6.3 | 19.0 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 6.2 | 18.5 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 6.1 | 24.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) response to kainic acid(GO:1904373) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 5.8 | 28.8 | GO:0030070 | insulin processing(GO:0030070) |
| 5.0 | 19.9 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) trophectodermal cell proliferation(GO:0001834) |
| 4.2 | 12.5 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 3.8 | 41.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 3.7 | 40.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 2.9 | 8.8 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 2.7 | 18.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 2.2 | 30.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 2.1 | 29.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 2.0 | 10.2 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 2.0 | 11.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 1.8 | 7.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 1.8 | 14.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 1.6 | 9.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.6 | 3.2 | GO:1904638 | response to resveratrol(GO:1904638) |
| 1.5 | 9.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 1.5 | 17.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.3 | 10.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 1.2 | 3.7 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 0.9 | 3.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.9 | 12.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.8 | 3.4 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.8 | 8.0 | GO:1903690 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.7 | 16.9 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.7 | 6.4 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.7 | 6.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.6 | 9.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.6 | 3.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.4 | 7.6 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.4 | 2.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.4 | 10.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.3 | 20.4 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.3 | 6.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 2.0 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.3 | 22.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 12.7 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.2 | 19.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 1.5 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.2 | 9.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 3.2 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.2 | 2.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 8.4 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 31.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 7.5 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 3.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 11.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 9.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 2.4 | GO:0003170 | heart valve development(GO:0003170) |
| 0.1 | 30.4 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 9.5 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.1 | 23.4 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 16.9 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 12.3 | GO:0016358 | dendrite development(GO:0016358) |
| 0.1 | 1.0 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.4 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.7 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 26.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 5.1 | 40.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 3.4 | 20.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 3.0 | 21.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 2.2 | 30.6 | GO:0031045 | dense core granule(GO:0031045) |
| 1.9 | 17.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.9 | 44.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.4 | 29.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.2 | 9.7 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 1.1 | 8.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.8 | 3.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.8 | 11.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.6 | 44.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.6 | 7.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.5 | 34.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.5 | 9.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 17.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 6.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 10.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 3.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 8.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 14.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 10.4 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 64.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 28.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 60.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.2 | 6.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 16.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 20.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 5.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 16.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 2.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 10.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 11.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 5.4 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 63.7 | GO:0097458 | neuron part(GO:0097458) |
| 0.0 | 2.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 8.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.4 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 5.9 | 29.3 | GO:1904315 | benzodiazepine receptor activity(GO:0008503) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 5.2 | 41.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 5.2 | 41.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 4.5 | 40.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 3.9 | 11.8 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 3.7 | 51.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 3.4 | 10.1 | GO:0030977 | taurine binding(GO:0030977) |
| 3.3 | 50.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.5 | 12.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.8 | 3.7 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 1.8 | 9.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.8 | 8.8 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 1.6 | 19.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.3 | 12.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.2 | 7.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 1.1 | 30.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 1.1 | 48.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 1.0 | 8.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.9 | 26.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.9 | 20.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.8 | 19.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.7 | 12.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.5 | 6.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 9.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.5 | 14.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 12.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 25.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 23.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.4 | 18.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 8.0 | GO:0002162 | dystroglycan binding(GO:0002162) microtubule plus-end binding(GO:0051010) |
| 0.3 | 1.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.3 | 2.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 26.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.3 | 16.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 3.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 3.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 3.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 9.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 28.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 8.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 6.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 1.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 19.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 7.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 14.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 10.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.4 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 3.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 6.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 41.0 | GO:0005215 | transporter activity(GO:0005215) |
| 0.0 | 6.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 8.0 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 31.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 1.1 | 24.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 21.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.5 | 26.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 9.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 3.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 9.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 8.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 8.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 10.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 6.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 15.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 10.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 41.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 2.1 | 55.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 1.7 | 29.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.9 | 10.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.7 | 11.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.6 | 11.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.6 | 8.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 8.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 31.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.3 | 9.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 19.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 3.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 12.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 9.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 3.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 10.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |