Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
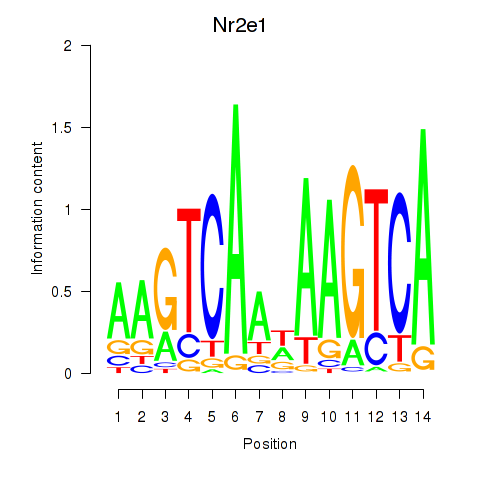
Results for Nr2e1
Z-value: 0.67
Transcription factors associated with Nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e1
|
ENSRNOG00000050550 | nuclear receptor subfamily 2, group E, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e1 | rn6_v1_chr20_-_47306318_47306318 | -0.34 | 4.9e-10 | Click! |
Activity profile of Nr2e1 motif
Sorted Z-values of Nr2e1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_176381477 | 42.67 |
ENSRNOT00000048367
|
Slco1a6
|
solute carrier organic anion transporter family, member 1a6 |
| chr4_-_176294997 | 35.69 |
ENSRNOT00000015112
ENSRNOT00000051461 ENSRNOT00000000026 ENSRNOT00000039877 ENSRNOT00000049303 |
Slc21a4
|
kidney specific organic anion transporter |
| chr17_-_43543172 | 30.20 |
ENSRNOT00000080684
ENSRNOT00000029626 ENSRNOT00000082719 |
Slc17a3
|
solute carrier family 17 member 3 |
| chr3_-_63568464 | 27.32 |
ENSRNOT00000068494
|
AABR07052585.2
|
|
| chr8_-_62948720 | 19.83 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr2_+_222214885 | 16.29 |
ENSRNOT00000055723
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr7_+_71572941 | 15.73 |
ENSRNOT00000007255
|
Sdc2
|
syndecan 2 |
| chr5_-_77492013 | 14.87 |
ENSRNOT00000012293
|
LOC259245
|
alpha-2u globulin PGCL5 |
| chr5_-_52767592 | 14.03 |
ENSRNOT00000048383
|
LOC298111
|
alpha2u globulin |
| chr2_+_198655437 | 13.57 |
ENSRNOT00000028781
|
Hfe2
|
hemochromatosis type 2 (juvenile) |
| chr8_+_103459161 | 12.94 |
ENSRNOT00000074047
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr2_-_210703606 | 12.82 |
ENSRNOT00000077563
|
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr2_+_153803349 | 11.95 |
ENSRNOT00000088565
|
Mme
|
membrane metallo-endopeptidase |
| chr2_-_210703265 | 11.75 |
ENSRNOT00000080228
|
Gstm6l
|
glutathione S-transferase, mu 6-like |
| chr1_+_13595295 | 10.40 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr5_-_79874671 | 9.08 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr16_+_2706428 | 8.83 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr18_-_64061472 | 7.57 |
ENSRNOT00000022519
|
Fam210a
|
family with sequence similarity 210, member A |
| chr1_+_152072665 | 7.55 |
ENSRNOT00000022538
|
Rab38
|
RAB38, member RAS oncogene family |
| chr4_+_32541706 | 7.34 |
ENSRNOT00000050827
|
Sdhaf3
|
succinate dehydrogenase complex assembly factor 3 |
| chr10_-_59883839 | 7.21 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr5_+_43603043 | 6.82 |
ENSRNOT00000009899
|
Epha7
|
Eph receptor A7 |
| chr13_+_89755845 | 5.60 |
ENSRNOT00000005550
|
Nectin4
|
nectin cell adhesion molecule 4 |
| chr8_-_22821397 | 5.52 |
ENSRNOT00000045488
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr1_+_203509731 | 5.48 |
ENSRNOT00000028007
|
Hmx3
|
H6 family homeobox 3 |
| chr17_-_89923423 | 4.36 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr10_+_55555089 | 4.32 |
ENSRNOT00000006597
|
Slc25a35
|
solute carrier family 25, member 35 |
| chr9_+_30939555 | 4.29 |
ENSRNOT00000016887
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr2_-_192960294 | 3.56 |
ENSRNOT00000012420
|
LOC102552326
|
late cornified envelope protein 5A-like |
| chr9_+_118332517 | 3.19 |
ENSRNOT00000031943
|
LOC103690198
|
taste receptor type 2 member 143 |
| chr14_-_107160334 | 3.17 |
ENSRNOT00000012131
|
Ehbp1
|
EH domain binding protein 1 |
| chr14_+_104191517 | 3.12 |
ENSRNOT00000006573
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chrX_-_106116886 | 3.01 |
ENSRNOT00000052009
|
Tcp11x2
|
t-complex 11 family, X-linked 2 |
| chr5_-_138462864 | 2.69 |
ENSRNOT00000011325
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr3_+_37146955 | 2.48 |
ENSRNOT00000030442
|
Tas2r134
|
taste receptor, type 2, member 134 |
| chr4_+_71790701 | 2.20 |
ENSRNOT00000085902
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chr8_+_117812099 | 1.72 |
ENSRNOT00000077228
|
Ccdc51
|
coiled-coil domain containing 51 |
| chr20_-_32628953 | 1.68 |
ENSRNOT00000000451
|
Gprc6a
|
G protein-coupled receptor, class C, group 6, member A |
| chr1_+_167758636 | 1.58 |
ENSRNOT00000024957
|
Olr46
|
olfactory receptor 46 |
| chr4_+_72332490 | 1.54 |
ENSRNOT00000088294
|
LOC103692138
|
olfactory receptor 6B1 |
| chr18_-_72649915 | 1.28 |
ENSRNOT00000072205
|
AABR07032582.1
|
|
| chr1_-_63457134 | 1.23 |
ENSRNOT00000083436
|
AABR07071874.1
|
|
| chr1_+_230011125 | 1.09 |
ENSRNOT00000087890
|
Olr352
|
olfactory receptor 352 |
| chr1_-_168160811 | 1.03 |
ENSRNOT00000020981
|
Olr70
|
olfactory receptor 70 |
| chr12_+_27155587 | 0.72 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr5_-_12199283 | 0.56 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr1_-_7335936 | 0.51 |
ENSRNOT00000020453
|
Ltv1
|
LTV1 ribosome biogenesis factor |
| chr17_+_63635086 | 0.08 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 35.7 | GO:0051958 | methotrexate transport(GO:0051958) |
| 5.4 | 16.3 | GO:0006214 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 3.0 | 12.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 2.7 | 30.2 | GO:0015747 | urate transport(GO:0015747) |
| 2.3 | 9.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 1.9 | 7.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 1.8 | 5.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.5 | 7.3 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 1.2 | 42.7 | GO:0042168 | heme metabolic process(GO:0042168) |
| 1.1 | 6.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.1 | 4.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.9 | 13.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.4 | 15.7 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.4 | 7.2 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 4.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.2 | 3.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 5.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 5.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 3.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 5.5 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 12.9 | GO:0010952 | positive regulation of peptidase activity(GO:0010952) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 8.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 19.8 | GO:0007155 | cell adhesion(GO:0007155) |
| 0.0 | 4.3 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 1.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 13.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.3 | 7.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.5 | 9.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 12.0 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 3.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 4.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 7.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 6.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 30.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 4.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 5.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 14.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 101.8 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 35.7 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 5.4 | 16.3 | GO:0004159 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 3.0 | 30.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 2.5 | 7.5 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 2.4 | 7.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 2.3 | 13.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 1.1 | 9.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.1 | 8.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.7 | 6.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.6 | 24.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.5 | 4.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.4 | 3.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 12.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 4.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 12.0 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.1 | 14.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 36.2 | GO:0005215 | transporter activity(GO:0005215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.3 | 9.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 13.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 6.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 8.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 7.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 16.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.8 | 15.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 13.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 12.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 9.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 8.8 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |