Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
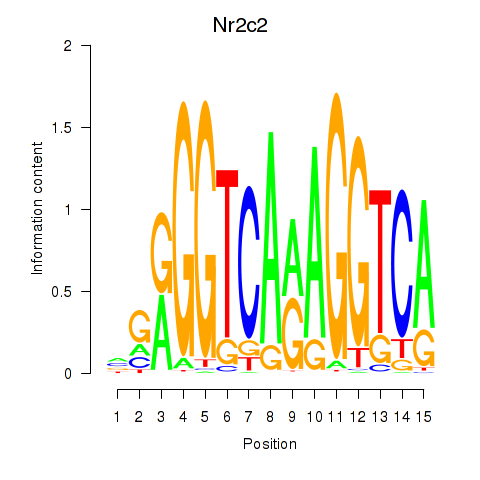
Results for Nr2c2
Z-value: 0.62
Transcription factors associated with Nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c2
|
ENSRNOG00000010536 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c2 | rn6_v1_chr4_+_124005280_124005280 | 0.49 | 1.9e-20 | Click! |
Activity profile of Nr2c2 motif
Sorted Z-values of Nr2c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_100231460 | 22.38 |
ENSRNOT00000032200
|
Acpt
|
acid phosphatase, testicular |
| chr9_+_110721322 | 19.86 |
ENSRNOT00000079170
|
AABR07068593.1
|
|
| chr2_+_78825074 | 15.81 |
ENSRNOT00000032027
|
March11
|
membrane associated ring-CH-type finger 11 |
| chr20_+_13778178 | 15.65 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr7_-_28715224 | 11.08 |
ENSRNOT00000065899
|
Parpbp
|
PARP1 binding protein |
| chr8_+_28075551 | 10.50 |
ENSRNOT00000012078
|
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chrX_-_123662350 | 9.88 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr10_-_97754445 | 9.70 |
ENSRNOT00000088215
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr3_+_171597241 | 9.65 |
ENSRNOT00000029558
|
LOC689618
|
similar to Protein C20orf85 homolog |
| chr13_+_51795867 | 9.43 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr17_+_42302540 | 9.26 |
ENSRNOT00000025389
|
Gmnn
|
geminin, DNA replication inhibitor |
| chr2_+_195719543 | 9.22 |
ENSRNOT00000028324
|
Celf3
|
CUGBP, Elav-like family member 3 |
| chr1_+_192233910 | 9.09 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr20_+_3351303 | 9.01 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr13_+_110511668 | 8.91 |
ENSRNOT00000006235
|
Nek2
|
NIMA-related kinase 2 |
| chr1_+_100086520 | 8.78 |
ENSRNOT00000025723
|
Klk1
|
kallikrein 1 |
| chr14_-_91556231 | 8.57 |
ENSRNOT00000039182
|
Zpbp
|
zona pellucida binding protein |
| chr7_-_12346475 | 8.43 |
ENSRNOT00000060708
|
Mum1
|
melanoma associated antigen (mutated) 1 |
| chr1_+_100224401 | 8.39 |
ENSRNOT00000025846
|
Fv1
|
Friend virus susceptibility 1 |
| chr16_-_24951612 | 8.24 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr16_+_20121352 | 8.16 |
ENSRNOT00000025347
|
Insl3
|
insulin-like 3 |
| chr7_-_82687130 | 7.67 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr18_-_16542165 | 7.56 |
ENSRNOT00000079381
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr9_+_81783349 | 7.54 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr8_+_117170620 | 7.42 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr7_-_136853957 | 7.36 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr10_+_11373346 | 7.27 |
ENSRNOT00000044624
|
LOC108348151
|
coronin-7-like |
| chr10_-_55965216 | 7.04 |
ENSRNOT00000057058
|
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr1_-_226791773 | 7.02 |
ENSRNOT00000082482
ENSRNOT00000065376 ENSRNOT00000054812 ENSRNOT00000086669 |
LOC100911215
|
T-cell surface glycoprotein CD5-like |
| chr3_+_110367939 | 6.90 |
ENSRNOT00000010406
|
Bub1b
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr3_-_160922341 | 6.86 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr1_-_163071508 | 6.83 |
ENSRNOT00000019053
|
Myo7a
|
myosin VIIA |
| chr6_-_138093643 | 6.67 |
ENSRNOT00000045874
|
Igh-6
|
immunoglobulin heavy chain 6 |
| chr8_+_127171537 | 6.60 |
ENSRNOT00000078625
ENSRNOT00000051290 ENSRNOT00000087753 |
Golga4
|
golgin A4 |
| chr18_+_68983545 | 6.56 |
ENSRNOT00000085317
|
Stard6
|
StAR-related lipid transfer domain containing 6 |
| chr11_+_61531571 | 6.49 |
ENSRNOT00000093467
ENSRNOT00000002727 |
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr10_+_31508512 | 6.46 |
ENSRNOT00000031580
|
Fam71b
|
family with sequence similarity 71, member B |
| chr5_-_49439358 | 6.43 |
ENSRNOT00000010909
|
Spaca1
|
sperm acrosome associated 1 |
| chr6_+_122254841 | 6.35 |
ENSRNOT00000005055
|
Gpr65
|
G-protein coupled receptor 65 |
| chr3_-_173947018 | 6.24 |
ENSRNOT00000078047
|
Sycp2
|
synaptonemal complex protein 2 |
| chr11_+_47188495 | 6.14 |
ENSRNOT00000002188
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr3_-_153454160 | 6.09 |
ENSRNOT00000010298
|
Ghrh
|
growth hormone releasing hormone |
| chr7_+_141355994 | 6.08 |
ENSRNOT00000081195
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr1_-_206282575 | 6.06 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr11_+_61531416 | 6.06 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chrX_+_74273182 | 6.05 |
ENSRNOT00000003922
|
Tsx
|
testis specific X-linked gene |
| chr2_-_207300854 | 5.95 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr6_-_26855658 | 5.83 |
ENSRNOT00000011635
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr17_+_55065621 | 5.80 |
ENSRNOT00000086342
|
Zfp438
|
zinc finger protein 438 |
| chr1_-_82952936 | 5.79 |
ENSRNOT00000075546
|
RGD1565655
|
similar to BC049730 protein |
| chr1_+_143583675 | 5.78 |
ENSRNOT00000026385
|
Fam103a1
|
family with sequence similarity 103, member A1 |
| chr8_-_62541959 | 5.68 |
ENSRNOT00000068108
|
Clk3
|
CDC-like kinase 3 |
| chr10_-_77512032 | 5.63 |
ENSRNOT00000003295
|
Pctp
|
phosphatidylcholine transfer protein |
| chr3_+_2462466 | 5.61 |
ENSRNOT00000014087
|
Rnf208
|
ring finger protein 208 |
| chr19_-_37675077 | 5.58 |
ENSRNOT00000031033
|
Enkd1
|
enkurin domain containing 1 |
| chr1_+_144069638 | 5.38 |
ENSRNOT00000026870
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr5_+_57947716 | 5.36 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr1_-_64350338 | 5.36 |
ENSRNOT00000078444
|
Cacng8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr12_-_2438817 | 5.35 |
ENSRNOT00000037059
|
Ccl25
|
C-C motif chemokine ligand 25 |
| chr9_-_10170428 | 5.34 |
ENSRNOT00000073048
|
LOC316124
|
similar to gonadotropin-regulated long chain acyl-CoA synthetase |
| chr10_+_55169282 | 5.32 |
ENSRNOT00000005423
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr13_+_48287873 | 5.26 |
ENSRNOT00000068223
|
Fam72a
|
family with sequence similarity 72, member A |
| chrX_+_156873849 | 5.24 |
ENSRNOT00000085410
|
Arhgap4
|
Rho GTPase activating protein 4 |
| chr1_-_166677026 | 5.21 |
ENSRNOT00000026644
ENSRNOT00000076714 |
Art2b
|
ADP-ribosyltransferase 2b |
| chr4_-_108008484 | 5.11 |
ENSRNOT00000007971
|
Ctnna2
|
catenin alpha 2 |
| chr1_+_166983175 | 5.05 |
ENSRNOT00000027028
|
Anapc15
|
anaphase promoting complex subunit 15 |
| chr10_-_56368360 | 5.01 |
ENSRNOT00000019674
|
Slc35g3
|
solute carrier family 35, member G3 |
| chr20_+_5619012 | 4.95 |
ENSRNOT00000000577
|
Ggnbp1
|
gametogenetin binding protein 1 |
| chr1_+_220423426 | 4.94 |
ENSRNOT00000072647
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr19_-_20508711 | 4.93 |
ENSRNOT00000032408
|
LOC680913
|
hypothetical protein LOC680913 |
| chr12_+_25036605 | 4.92 |
ENSRNOT00000001996
ENSRNOT00000084427 |
Limk1
|
LIM domain kinase 1 |
| chr4_+_157659147 | 4.91 |
ENSRNOT00000048379
|
Iffo1
|
intermediate filament family orphan 1 |
| chr12_+_19680712 | 4.84 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr12_-_38504774 | 4.82 |
ENSRNOT00000011286
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr3_-_80000091 | 4.82 |
ENSRNOT00000079394
|
Madd
|
MAP-kinase activating death domain |
| chr14_+_84465515 | 4.79 |
ENSRNOT00000035591
|
Osm
|
oncostatin M |
| chr16_+_75572070 | 4.77 |
ENSRNOT00000043486
|
Defb52
|
defensin beta 52 |
| chr1_-_80311043 | 4.75 |
ENSRNOT00000068092
|
Klc3
|
kinesin light chain 3 |
| chr1_-_89045586 | 4.74 |
ENSRNOT00000063808
|
Zbtb32
|
zinc finger and BTB domain containing 32 |
| chr1_-_262014066 | 4.74 |
ENSRNOT00000087083
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr14_+_8182383 | 4.72 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr6_-_128690741 | 4.71 |
ENSRNOT00000035826
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr2_+_235596907 | 4.63 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr3_-_16999720 | 4.61 |
ENSRNOT00000074382
|
RGD1563231
|
similar to immunoglobulin kappa-chain VK-1 |
| chr1_+_229642412 | 4.61 |
ENSRNOT00000017109
|
Lpxn
|
leupaxin |
| chr20_-_5927070 | 4.60 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr4_+_30807879 | 4.59 |
ENSRNOT00000013184
|
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr7_-_12777901 | 4.59 |
ENSRNOT00000017245
|
Tmem259
|
transmembrane protein 259 |
| chr10_-_14937336 | 4.56 |
ENSRNOT00000025494
|
Sox8
|
SRY box 8 |
| chr1_-_221041401 | 4.50 |
ENSRNOT00000064136
|
Pcnx3
|
pecanex homolog 3 (Drosophila) |
| chr16_-_59366824 | 4.49 |
ENSRNOT00000015062
|
RGD1304810
|
similar to 6430573F11Rik protein |
| chr14_+_85230648 | 4.47 |
ENSRNOT00000089866
|
Ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr6_+_43001948 | 4.43 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr14_-_108284619 | 4.41 |
ENSRNOT00000078814
|
LOC690352
|
hypothetical protein LOC690352 |
| chr7_-_26690323 | 4.37 |
ENSRNOT00000087536
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr5_-_122828398 | 4.35 |
ENSRNOT00000030112
|
RGD1562532
|
hypothetical gene supported by BC079057 |
| chr5_+_152333043 | 4.34 |
ENSRNOT00000021342
|
Ubxn11
|
UBX domain protein 11 |
| chr3_+_161272385 | 4.33 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chrX_+_19270181 | 4.32 |
ENSRNOT00000029397
|
Usp51
|
ubiquitin specific peptidase 51 |
| chr4_+_162437089 | 4.31 |
ENSRNOT00000038801
|
Clec2d2
|
C-type lectin domain family 2 member D2 |
| chr2_+_123791335 | 4.30 |
ENSRNOT00000077096
|
Adad1
|
adenosine deaminase domain containing 1 |
| chr1_-_187779675 | 4.22 |
ENSRNOT00000024648
|
Arl6ip1
|
ADP-ribosylation factor like GTPase 6 interacting protein 1 |
| chr11_+_54137639 | 4.22 |
ENSRNOT00000066343
|
LOC100909977
|
leukocyte surface antigen CD47-like |
| chr1_+_265904566 | 4.20 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr4_-_98735346 | 4.20 |
ENSRNOT00000008473
|
Tex37
|
testis expressed 37 |
| chr2_+_187218851 | 4.20 |
ENSRNOT00000017798
|
Sh2d2a
|
SH2 domain containing 2A |
| chr5_-_110706656 | 4.19 |
ENSRNOT00000033781
|
Izumo3
|
IZUMO family member 3 |
| chr3_-_160738927 | 4.18 |
ENSRNOT00000043470
|
Slpil2
|
antileukoproteinase-like 2 |
| chr8_+_44001096 | 4.15 |
ENSRNOT00000084164
|
Tmem225
|
transmembrane protein 225 |
| chr12_+_22026075 | 4.15 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr14_-_108285008 | 4.14 |
ENSRNOT00000087501
|
LOC690352
|
hypothetical protein LOC690352 |
| chr7_+_3173287 | 4.13 |
ENSRNOT00000092037
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr16_-_10529061 | 4.09 |
ENSRNOT00000086215
|
Gprin2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr12_+_37709860 | 4.08 |
ENSRNOT00000086244
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr20_+_4530342 | 4.05 |
ENSRNOT00000076352
ENSRNOT00000000478 ENSRNOT00000075925 |
Nelfe
|
negative elongation factor complex member E |
| chr12_-_47653638 | 4.02 |
ENSRNOT00000001577
|
Tchp
|
trichoplein, keratin filament binding |
| chr10_+_47961056 | 4.02 |
ENSRNOT00000027312
|
Fam83g
|
family with sequence similarity 83, member G |
| chr5_+_79382096 | 4.01 |
ENSRNOT00000085461
|
Tmem268
|
transmembrane protein 268 |
| chr18_-_410098 | 4.01 |
ENSRNOT00000084138
|
LOC102546764
|
cx9C motif-containing protein 4-like |
| chr4_+_88832178 | 4.00 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr10_+_30038709 | 4.00 |
ENSRNOT00000005898
|
Il12b
|
interleukin 12B |
| chr2_-_29248174 | 4.00 |
ENSRNOT00000071167
|
LOC684444
|
similar to HIStone family member (his-41) |
| chr17_+_44556039 | 4.00 |
ENSRNOT00000086540
|
Prss16
|
protease, serine 16 |
| chr14_+_85814824 | 3.98 |
ENSRNOT00000090046
ENSRNOT00000083756 |
Ankrd36
|
ankyrin repeat domain 36 |
| chrX_+_15273933 | 3.96 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr1_-_224986291 | 3.96 |
ENSRNOT00000026284
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr5_+_137257287 | 3.94 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr7_+_80351774 | 3.93 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
| chr3_-_7795758 | 3.93 |
ENSRNOT00000045919
|
Ntng2
|
netrin G2 |
| chr12_+_17614632 | 3.92 |
ENSRNOT00000031896
|
Prkar1b
|
protein kinase cAMP-dependent type 1 regulatory subunit beta |
| chr18_+_45023932 | 3.89 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr17_-_14949064 | 3.88 |
ENSRNOT00000079489
ENSRNOT00000081454 |
AABR07027128.1
|
|
| chr15_+_90364605 | 3.88 |
ENSRNOT00000020876
|
Trim52
|
tripartite motif-containing 52 |
| chr8_-_6076598 | 3.84 |
ENSRNOT00000090774
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr7_-_62162453 | 3.83 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr3_-_11102515 | 3.82 |
ENSRNOT00000035580
|
Fam78a
|
family with sequence similarity 78, member A |
| chr6_-_39363367 | 3.78 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr19_+_49637016 | 3.73 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
| chr20_+_5535432 | 3.72 |
ENSRNOT00000040859
|
Syngap1
|
synaptic Ras GTPase activating protein 1 |
| chr3_-_1584946 | 3.71 |
ENSRNOT00000031058
|
Pax8
|
paired box 8 |
| chr19_+_37330930 | 3.70 |
ENSRNOT00000022439
|
Plekhg4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chr5_+_103479767 | 3.69 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr8_+_123126667 | 3.69 |
ENSRNOT00000015643
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr3_-_160739137 | 3.68 |
ENSRNOT00000075836
|
Slpil2
|
antileukoproteinase-like 2 |
| chrX_+_14019961 | 3.65 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr17_+_15762030 | 3.62 |
ENSRNOT00000089310
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr1_-_36112993 | 3.61 |
ENSRNOT00000023161
|
Med10
|
mediator complex subunit 10 |
| chr3_+_72385666 | 3.58 |
ENSRNOT00000011168
|
Prg2
|
proteoglycan 2 |
| chr11_+_67082193 | 3.57 |
ENSRNOT00000003129
|
Cd86
|
CD86 molecule |
| chr5_-_139931366 | 3.56 |
ENSRNOT00000015250
|
Smap2
|
small ArfGAP2 |
| chr13_-_93746994 | 3.56 |
ENSRNOT00000005072
|
Opn3
|
opsin 3 |
| chr10_-_75202030 | 3.51 |
ENSRNOT00000050349
|
Olr1522
|
olfactory receptor 1522 |
| chr10_+_104437648 | 3.49 |
ENSRNOT00000035001
|
AC130970.1
|
|
| chr4_+_170958196 | 3.48 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr8_-_80631873 | 3.47 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr6_-_132608600 | 3.45 |
ENSRNOT00000015855
|
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr3_-_4405194 | 3.44 |
ENSRNOT00000072025
|
AABR07051266.1
|
|
| chr9_-_81565416 | 3.44 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr5_-_152446775 | 3.43 |
ENSRNOT00000021919
|
Catsper4
|
cation channel, sperm associated 4 |
| chr18_-_68983619 | 3.43 |
ENSRNOT00000014670
|
LOC361346
|
similar to chromosome 18 open reading frame 54 |
| chr20_-_4530126 | 3.42 |
ENSRNOT00000000481
|
Skiv2l
|
Ski2 like RNA helicase |
| chr16_-_71040847 | 3.38 |
ENSRNOT00000020606
|
Star
|
steroidogenic acute regulatory protein |
| chr9_+_76440186 | 3.37 |
ENSRNOT00000049479
|
RGD1562431
|
similar to chromosome 20 open reading frame 81 |
| chr7_-_70556827 | 3.34 |
ENSRNOT00000007721
|
Kif5a
|
kinesin family member 5A |
| chr8_+_55603968 | 3.31 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr10_+_11090314 | 3.29 |
ENSRNOT00000086305
|
Coro7
|
coronin 7 |
| chr16_-_20097287 | 3.23 |
ENSRNOT00000025162
|
Unc13a
|
unc-13 homolog A |
| chr7_-_144960527 | 3.22 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr5_-_28737719 | 3.22 |
ENSRNOT00000009718
|
Necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr1_-_142020525 | 3.20 |
ENSRNOT00000042558
|
Cib1
|
calcium and integrin binding 1 |
| chr20_+_2501252 | 3.20 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr1_-_81946714 | 3.18 |
ENSRNOT00000027578
|
Grik5
|
glutamate ionotropic receptor kainate type subunit 5 |
| chr3_+_176494768 | 3.17 |
ENSRNOT00000040270
|
Col20a1
|
collagen type XX alpha 1 chain |
| chr1_-_82610350 | 3.16 |
ENSRNOT00000028177
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1 |
| chr7_-_70969905 | 3.12 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr17_-_49990982 | 3.11 |
ENSRNOT00000018458
ENSRNOT00000080628 |
Mplkip
|
M-phase specific PLK1 interacting protein |
| chr10_-_7029136 | 3.07 |
ENSRNOT00000091942
|
AC129395.1
|
|
| chr2_+_178117466 | 3.07 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr2_-_30780121 | 3.06 |
ENSRNOT00000025129
|
Cenph
|
centromere protein H |
| chrX_-_123980357 | 3.04 |
ENSRNOT00000049435
|
Rhox8
|
reproductive homeobox 8 |
| chr16_+_20109200 | 3.04 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr3_+_155269347 | 3.04 |
ENSRNOT00000021308
|
Fam83d
|
family with sequence similarity 83, member D |
| chr7_-_117605141 | 3.02 |
ENSRNOT00000035199
|
Fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr1_+_212558257 | 3.01 |
ENSRNOT00000024912
|
Prap1
|
proline-rich acidic protein 1 |
| chr1_+_97712177 | 3.00 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr13_-_78608844 | 2.98 |
ENSRNOT00000003671
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr12_+_47193964 | 2.95 |
ENSRNOT00000001552
ENSRNOT00000039281 |
Cabp1
|
calcium binding protein 1 |
| chr7_+_12471824 | 2.95 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chr12_-_24537313 | 2.95 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr12_+_39679688 | 2.94 |
ENSRNOT00000050398
|
Fam216a
|
family with sequence similarity 216, member A |
| chr14_+_100135271 | 2.94 |
ENSRNOT00000032965
|
Fbxo48
|
F-box protein 48 |
| chr7_-_121302740 | 2.94 |
ENSRNOT00000067032
|
Rpl3
|
ribosomal protein L3 |
| chr3_-_168033457 | 2.92 |
ENSRNOT00000055111
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr8_+_70112925 | 2.92 |
ENSRNOT00000082401
|
Megf11
|
multiple EGF-like-domains 11 |
| chr17_+_27452638 | 2.91 |
ENSRNOT00000038349
|
Cage1
|
cancer antigen 1 |
| chr5_+_9279970 | 2.89 |
ENSRNOT00000067977
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr9_-_50868238 | 2.89 |
ENSRNOT00000015600
|
Tex30
|
testis expressed 30 |
| chr20_-_29990111 | 2.87 |
ENSRNOT00000048029
|
Cdh23
|
cadherin-related 23 |
| chr4_+_57204944 | 2.84 |
ENSRNOT00000085027
|
Strip2
|
striatin interacting protein 2 |
| chr1_-_47307488 | 2.84 |
ENSRNOT00000090033
|
Ezr
|
ezrin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 3.0 | 9.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.0 | 9.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 3.0 | 8.9 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 2.4 | 9.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.8 | 5.5 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 1.7 | 5.2 | GO:0019677 | NAD catabolic process(GO:0019677) |
| 1.6 | 4.8 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 1.5 | 7.5 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 1.4 | 4.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 1.4 | 6.9 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.4 | 5.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.4 | 10.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.3 | 4.0 | GO:1904612 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 1.3 | 9.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.3 | 3.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.3 | 3.8 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 1.2 | 3.6 | GO:0002215 | defense response to nematode(GO:0002215) |
| 1.2 | 3.6 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 1.2 | 6.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.2 | 3.5 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 1.1 | 4.6 | GO:0072289 | metanephric nephron tubule formation(GO:0072289) |
| 1.1 | 3.4 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 1.1 | 3.3 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 1.1 | 11.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 1.1 | 5.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.1 | 3.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.0 | 3.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.0 | 6.2 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 1.0 | 9.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 1.0 | 6.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 1.0 | 3.0 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 1.0 | 4.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 1.0 | 4.0 | GO:0030221 | basophil differentiation(GO:0030221) |
| 1.0 | 3.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 1.0 | 4.8 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.9 | 2.6 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.8 | 5.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.8 | 14.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.8 | 2.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.8 | 8.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.8 | 10.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.8 | 3.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.8 | 5.5 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.8 | 2.3 | GO:0046671 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.8 | 2.3 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.7 | 2.2 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 0.7 | 2.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.7 | 2.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.7 | 2.2 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.7 | 5.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.7 | 2.2 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.7 | 3.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.7 | 5.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.6 | 5.8 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.6 | 3.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.6 | 8.2 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.6 | 9.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.6 | 1.8 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.6 | 1.2 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.6 | 4.7 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.6 | 4.6 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.6 | 2.3 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.6 | 5.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.6 | 1.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.5 | 4.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.5 | 4.9 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.5 | 3.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.5 | 1.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.5 | 3.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.5 | 2.5 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.5 | 2.0 | GO:0019474 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.5 | 1.5 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.5 | 1.5 | GO:0002780 | antimicrobial peptide biosynthetic process(GO:0002777) antibacterial peptide biosynthetic process(GO:0002780) detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.5 | 2.0 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.5 | 4.4 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.5 | 1.4 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.5 | 3.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.5 | 1.8 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.5 | 4.1 | GO:1900364 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 4.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.4 | 1.3 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.4 | 1.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.4 | 4.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 5.1 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.4 | 1.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.4 | 13.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.4 | 1.6 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 4.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.4 | 1.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 | 4.8 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.4 | 3.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.4 | 3.8 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.4 | 3.4 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.4 | 2.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.4 | 1.9 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.4 | 0.7 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.4 | 1.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.4 | 2.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.4 | 3.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 2.0 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.3 | 4.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.3 | 8.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 2.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 1.6 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.3 | 1.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.3 | 1.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.3 | 2.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 2.7 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.3 | 5.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 0.9 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.3 | 1.7 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.3 | 1.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.3 | 2.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.7 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.3 | 0.8 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.3 | 4.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.3 | 3.0 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 0.3 | 6.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.3 | 0.5 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.3 | 8.0 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.3 | 4.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 1.3 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 1.6 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 0.5 | GO:0034163 | negative regulation of interferon-alpha production(GO:0032687) regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.3 | 3.1 | GO:0051383 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.2 | 1.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.2 | 1.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.2 | 1.2 | GO:1903203 | regulation of oxidative stress-induced neuron death(GO:1903203) negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.2 | 0.7 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.2 | 2.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 3.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 1.6 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 0.7 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 4.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 4.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.9 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.2 | 3.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 6.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.2 | 2.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 0.6 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.2 | 5.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 1.2 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.2 | 0.6 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 2.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 2.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 3.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.2 | 2.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.2 | 2.6 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.2 | 0.4 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 0.7 | GO:0015746 | citrate transport(GO:0015746) |
| 0.2 | 1.7 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 1.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 0.6 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.2 | 2.3 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 2.0 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 2.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 2.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 1.4 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 1.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 2.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 3.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 15.6 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 1.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.9 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 3.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 4.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.7 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 2.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 2.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 17.0 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 2.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 5.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 2.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.8 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.5 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 1.9 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 2.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.1 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.0 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 3.0 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 3.1 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.1 | 1.0 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.5 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.1 | 3.7 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 1.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.2 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 | 0.7 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.8 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 1.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 2.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 1.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 2.1 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 0.6 | GO:0090662 | ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 1.2 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 1.7 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 2.0 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 8.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.7 | GO:0090042 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 1.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 2.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.5 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 2.2 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 2.1 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.2 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.1 | 3.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 1.4 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.9 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 2.5 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.5 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.7 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 0.5 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.2 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.4 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 3.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 1.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.8 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 2.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 2.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 4.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 1.4 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 1.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.0 | GO:0090314 | positive regulation of protein targeting to membrane(GO:0090314) |
| 0.0 | 0.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.6 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 2.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.5 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 4.3 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 1.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 2.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.4 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 1.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 2.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 3.7 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 1.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.8 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.1 | GO:0010266 | response to vitamin B1(GO:0010266) |
| 0.0 | 7.6 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 3.5 | GO:0042098 | T cell proliferation(GO:0042098) |
| 0.0 | 1.0 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 6.5 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.1 | GO:0030509 | BMP signaling pathway(GO:0030509) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.9 | GO:0032173 | septin collar(GO:0032173) |
| 2.6 | 10.5 | GO:0042585 | germinal vesicle(GO:0042585) |
| 2.3 | 9.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 1.7 | 6.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.5 | 4.6 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 1.5 | 7.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 1.2 | 5.8 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 1.1 | 13.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.1 | 3.4 | GO:0055087 | Ski complex(GO:0055087) |
| 1.0 | 4.0 | GO:0032021 | NELF complex(GO:0032021) |
| 1.0 | 9.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.9 | 5.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.8 | 4.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.8 | 6.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.8 | 2.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.7 | 2.8 | GO:0044393 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 0.7 | 9.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.7 | 4.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.7 | 2.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.6 | 3.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.6 | 1.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.6 | 8.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.5 | 7.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.5 | 2.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 6.9 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.5 | 3.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 3.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.5 | 3.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.5 | 8.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 1.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 3.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 5.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 10.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 2.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.4 | 4.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 5.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.4 | 5.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 7.0 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 0.7 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.3 | 6.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 6.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 2.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.3 | 1.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 2.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 1.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.3 | 3.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 5.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 2.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 3.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 3.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 1.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 0.9 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 1.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 1.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 1.5 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 4.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 5.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 6.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 6.1 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 0.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 3.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 2.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 2.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 12.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 14.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 2.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 3.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 8.9 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 1.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 4.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 9.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 19.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 3.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 6.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 4.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 2.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.5 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 5.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 11.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 4.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 9.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 6.1 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 6.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 2.0 | 6.1 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 1.8 | 5.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.3 | 9.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.3 | 9.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.2 | 4.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.2 | 3.5 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 1.1 | 4.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 1.1 | 21.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 1.1 | 4.4 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.9 | 2.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.8 | 4.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.8 | 3.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.8 | 2.3 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.7 | 6.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.7 | 2.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.7 | 2.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.7 | 3.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.7 | 2.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.7 | 4.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.7 | 5.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.6 | 5.6 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.6 | 1.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.6 | 12.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.6 | 1.8 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.6 | 1.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.6 | 3.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.6 | 6.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.6 | 5.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 1.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.5 | 2.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.5 | 6.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.5 | 4.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 1.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.5 | 3.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.4 | 1.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.4 | 1.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.4 | 1.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 1.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.4 | 3.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.4 | 1.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.4 | 2.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 1.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.4 | 4.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.4 | 11.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.4 | 1.4 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.3 | 1.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.3 | 3.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 2.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 2.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.3 | 8.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 4.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 2.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 2.0 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 2.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 15.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 0.7 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.2 | 3.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 0.7 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 2.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 3.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.7 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.7 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 0.9 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.2 | 1.5 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 1.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 2.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 5.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 8.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 0.9 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 1.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 6.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 1.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 4.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 4.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 4.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 10.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 11.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 1.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 2.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.2 | 0.7 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.2 | 0.5 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.2 | 4.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 3.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 6.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 6.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 3.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 2.1 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.1 | 12.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.7 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 3.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.4 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 13.5 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 3.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.4 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 2.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 11.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 5.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 5.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 3.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.5 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.1 | 6.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 3.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.1 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 3.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 5.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 3.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.4 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.1 | 0.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 5.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 10.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 7.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 3.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 5.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.2 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0030911 | signal recognition particle binding(GO:0005047) TPR domain binding(GO:0030911) |
| 0.0 | 2.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 3.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 2.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 14.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 2.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 12.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 2.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 6.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 10.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 4.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 9.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 7.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 3.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 6.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 2.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 4.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 5.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 5.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.6 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 15.8 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.7 | 4.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.6 | 4.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.6 | 9.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.5 | 9.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.5 | 13.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 9.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.5 | 8.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 3.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 4.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 5.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.3 | 3.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 5.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 9.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.3 | 4.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 5.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 3.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 5.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 3.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 8.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 3.9 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.2 | 6.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 2.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 2.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 3.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 2.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 2.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 3.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 5.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 2.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 8.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 4.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 6.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 3.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 6.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 7.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 4.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.1 | 1.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.2 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 5.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 2.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.0 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 5.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 4.4 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |