Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
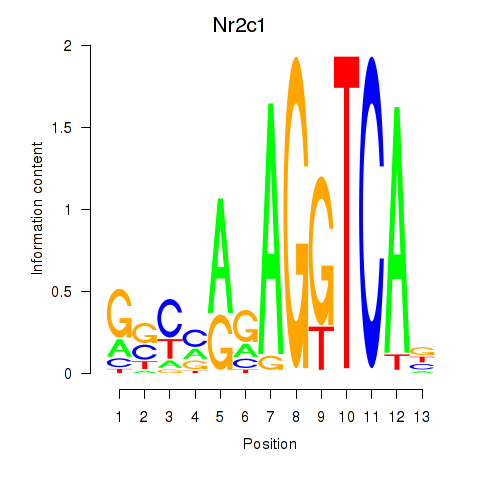
Results for Nr2c1
Z-value: 0.81
Transcription factors associated with Nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c1
|
ENSRNOG00000006983 | nuclear receptor subfamily 2, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c1 | rn6_v1_chr7_+_35070284_35070284 | -0.35 | 1.7e-10 | Click! |
Activity profile of Nr2c1 motif
Sorted Z-values of Nr2c1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_11240138 | 47.39 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr6_+_137184820 | 39.54 |
ENSRNOT00000073796
|
Adssl1
|
adenylosuccinate synthase like 1 |
| chr19_-_56677084 | 34.54 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr8_-_84506328 | 28.27 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr5_-_57372239 | 25.28 |
ENSRNOT00000012975
|
Aqp7
|
aquaporin 7 |
| chr1_-_221917901 | 24.36 |
ENSRNOT00000092270
|
Slc22a12
|
solute carrier family 22 member 12 |
| chr9_-_115850634 | 23.90 |
ENSRNOT00000049720
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr10_+_86337728 | 23.64 |
ENSRNOT00000085408
|
Tcap
|
titin-cap |
| chr10_+_71159869 | 20.82 |
ENSRNOT00000075977
ENSRNOT00000047427 |
Hnf1b
|
HNF1 homeobox B |
| chr9_+_65478496 | 20.82 |
ENSRNOT00000016060
|
Ndufb3
|
NADH:ubiquinone oxidoreductase subunit B3 |
| chr2_+_233615739 | 19.09 |
ENSRNOT00000051009
|
Pitx2
|
paired-like homeodomain 2 |
| chr6_+_8284878 | 16.97 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr1_+_31264755 | 16.11 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr5_-_101138427 | 14.04 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr5_+_173640780 | 13.83 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr1_-_222178725 | 12.40 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr1_+_101517714 | 12.38 |
ENSRNOT00000028423
|
Plekha4
|
pleckstrin homology domain containing A4 |
| chr10_-_50574539 | 12.35 |
ENSRNOT00000034261
|
Cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr4_+_102426224 | 11.87 |
ENSRNOT00000073768
|
LOC103694107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 pseudogene |
| chr8_-_55087832 | 11.78 |
ENSRNOT00000032152
|
Dlat
|
dihydrolipoamide S-acetyltransferase |
| chr7_-_117267803 | 11.33 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr14_-_83741969 | 10.89 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr7_-_117268300 | 10.39 |
ENSRNOT00000091840
|
Plec
|
plectin |
| chr1_+_175586097 | 10.09 |
ENSRNOT00000024933
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr7_-_144272578 | 9.82 |
ENSRNOT00000020676
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr3_-_48536235 | 9.11 |
ENSRNOT00000083536
|
Fap
|
fibroblast activation protein, alpha |
| chr9_+_65763437 | 8.88 |
ENSRNOT00000043558
|
Stradb
|
STE20-related kinase adaptor beta |
| chr2_+_198965685 | 8.82 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr9_-_9985630 | 8.54 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr19_-_55284663 | 8.51 |
ENSRNOT00000018387
|
Snai3
|
snail family transcriptional repressor 3 |
| chr14_-_17582823 | 8.41 |
ENSRNOT00000087686
|
LOC100911949
|
uncharacterized LOC100911949 |
| chr1_-_141579871 | 8.00 |
ENSRNOT00000020002
|
Anpep
|
alanyl aminopeptidase, membrane |
| chr1_+_222519615 | 7.81 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr2_+_95045034 | 7.52 |
ENSRNOT00000081305
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr15_+_108960562 | 7.41 |
ENSRNOT00000081906
ENSRNOT00000077482 |
Pcca
|
propionyl-CoA carboxylase alpha subunit |
| chr9_-_65763273 | 7.38 |
ENSRNOT00000015034
|
Trak2
|
trafficking kinesin protein 2 |
| chr15_+_19603288 | 7.18 |
ENSRNOT00000035491
|
LOC102552640
|
REST corepressor 2-like |
| chr7_+_120380544 | 6.74 |
ENSRNOT00000015009
|
Polr2f
|
RNA polymerase II subunit F |
| chr6_-_106971250 | 6.63 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr2_-_178389608 | 6.23 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr9_+_17105151 | 5.90 |
ENSRNOT00000091113
|
Tjap1
|
tight junction associated protein 1 |
| chr20_+_27954433 | 5.48 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr5_-_58476251 | 5.24 |
ENSRNOT00000013151
|
Stoml2
|
stomatin like 2 |
| chr1_+_212558257 | 4.16 |
ENSRNOT00000024912
|
Prap1
|
proline-rich acidic protein 1 |
| chr12_+_47407811 | 3.86 |
ENSRNOT00000001565
|
Hnf1a
|
HNF1 homeobox A |
| chr7_-_127081704 | 3.76 |
ENSRNOT00000055893
|
LOC108351584
|
uncharacterized LOC108351584 |
| chr6_-_21135880 | 3.65 |
ENSRNOT00000051239
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr20_-_3819200 | 3.60 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr8_+_127171537 | 3.24 |
ENSRNOT00000078625
ENSRNOT00000051290 ENSRNOT00000087753 |
Golga4
|
golgin A4 |
| chr11_-_61530830 | 3.05 |
ENSRNOT00000059666
ENSRNOT00000068345 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr8_-_43920420 | 2.77 |
ENSRNOT00000081179
|
Olr1339
|
olfactory receptor 1339 |
| chr3_+_37545238 | 2.74 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr2_+_148722231 | 2.45 |
ENSRNOT00000018022
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr3_-_148428494 | 2.29 |
ENSRNOT00000011350
|
Dusp15
|
dual specificity phosphatase 15 |
| chr7_-_120380200 | 2.08 |
ENSRNOT00000014878
|
RGD1359634
|
similar to RIKEN cDNA 1700088E04 |
| chr12_-_29952196 | 1.86 |
ENSRNOT00000086175
|
Tmem248
|
transmembrane protein 248 |
| chr10_-_87541851 | 1.77 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chr13_-_109770199 | 1.39 |
ENSRNOT00000043672
|
Fam71a
|
family with sequence similarity 71, member A |
| chr15_-_28446785 | 1.21 |
ENSRNOT00000015439
|
Olr1638
|
olfactory receptor 1638 |
| chr3_-_148428288 | 1.18 |
ENSRNOT00000072663
|
Dusp15
|
dual specificity phosphatase 15 |
| chr3_+_97723901 | 1.12 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr12_+_6283288 | 1.11 |
ENSRNOT00000001203
|
Wdr95
|
WD40 repeat domain 95 |
| chr1_+_85162452 | 1.07 |
ENSRNOT00000093347
|
Pak4
|
p21 (RAC1) activated kinase 4 |
| chr4_+_157181795 | 1.02 |
ENSRNOT00000017090
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr10_-_75202030 | 0.73 |
ENSRNOT00000050349
|
Olr1522
|
olfactory receptor 1522 |
| chr10_+_4411766 | 0.71 |
ENSRNOT00000003213
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr15_+_38069320 | 0.61 |
ENSRNOT00000014397
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr10_+_4313100 | 0.28 |
ENSRNOT00000074487
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr11_-_84037938 | 0.27 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr13_+_99004942 | 0.21 |
ENSRNOT00000085390
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr19_+_22569999 | 0.03 |
ENSRNOT00000022573
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.5 | 58.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 13.2 | 39.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 11.8 | 47.4 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 6.9 | 20.8 | GO:0061215 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 6.4 | 19.1 | GO:0021539 | subthalamus development(GO:0021539) pulmonary vein morphogenesis(GO:0060577) |
| 5.1 | 25.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 3.9 | 3.9 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 3.0 | 9.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 2.5 | 7.4 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 2.2 | 24.4 | GO:0015747 | urate transport(GO:0015747) |
| 2.2 | 8.8 | GO:0015879 | carnitine transport(GO:0015879) |
| 1.7 | 10.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.3 | 5.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.1 | 21.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.0 | 28.3 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.8 | 11.8 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.7 | 12.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.7 | 5.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.7 | 8.0 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.6 | 10.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.6 | 14.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.6 | 6.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.6 | 13.8 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.4 | 2.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.4 | 3.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 9.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 12.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.3 | 3.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 20.8 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.2 | 3.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 8.9 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 6.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 17.0 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 2.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 4.3 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 3.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.0 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 | 11.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 34.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 1.3 | 9.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.2 | 11.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.1 | 21.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 1.1 | 16.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.8 | 75.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.8 | 9.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.6 | 8.0 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 6.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.4 | 6.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.4 | 3.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 20.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 12.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.3 | 5.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 2.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 7.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 3.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 7.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 6.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 10.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 20.8 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 8.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 64.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 23.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 10.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 14.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 5.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 11.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 6.6 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 25.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 4.7 | 23.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 3.6 | 10.9 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 2.5 | 10.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 2.4 | 24.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 1.5 | 12.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 1.5 | 11.8 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 1.2 | 3.6 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 1.0 | 6.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.9 | 7.4 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.9 | 9.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.8 | 6.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.7 | 5.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.7 | 39.5 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.6 | 21.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.5 | 9.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 7.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.4 | 19.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 8.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 25.9 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.3 | 1.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 3.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 20.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 15.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 45.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 3.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 4.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 10.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 3.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 8.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 7.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 5.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 17.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 34.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.6 | 19.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.5 | 24.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 8.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 5.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 14.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 39.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 2.2 | 24.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 2.1 | 25.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.8 | 11.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 18.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 24.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.6 | 10.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 23.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 8.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.5 | 7.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 6.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.4 | 17.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 27.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 5.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 10.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 7.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 12.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 3.7 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |