Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
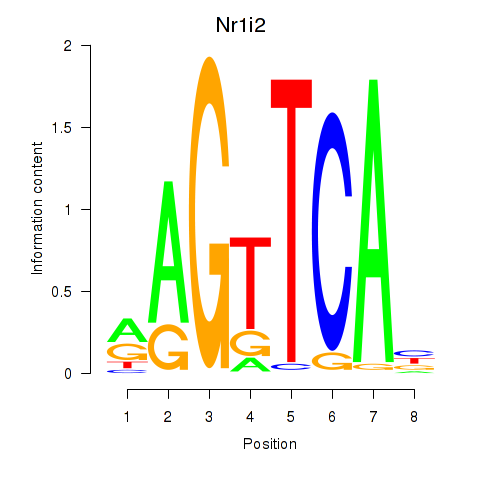
Results for Nr1i2
Z-value: 1.54
Transcription factors associated with Nr1i2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1i2
|
ENSRNOG00000002906 | nuclear receptor subfamily 1, group I, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1i2 | rn6_v1_chr11_+_65022100_65022100 | 0.19 | 5.4e-04 | Click! |
Activity profile of Nr1i2 motif
Sorted Z-values of Nr1i2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_22744407 | 61.64 |
ENSRNOT00000073710
|
Cmya5
|
cardiomyopathy associated 5 |
| chr15_+_33606124 | 59.34 |
ENSRNOT00000065210
|
AC115371.1
|
|
| chr10_+_11240138 | 41.88 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr8_+_26652121 | 41.76 |
ENSRNOT00000009342
|
Eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr17_-_18590536 | 41.18 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr19_-_56677084 | 40.49 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr8_-_84506328 | 39.95 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr1_+_72882806 | 39.78 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr2_-_200003443 | 39.43 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr14_+_26662965 | 37.81 |
ENSRNOT00000002621
|
Tecrl
|
trans-2,3-enoyl-CoA reductase-like |
| chrX_+_159158194 | 33.22 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr1_-_277181345 | 32.57 |
ENSRNOT00000038017
ENSRNOT00000038038 |
Nrap
|
nebulin-related anchoring protein |
| chr19_+_37235001 | 31.83 |
ENSRNOT00000020908
|
Nol3
|
nucleolar protein 3 |
| chr5_-_78985990 | 31.63 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr4_+_51553454 | 28.97 |
ENSRNOT00000073466
|
LOC100909784
|
leiomodin-2-like |
| chr4_+_99063181 | 28.84 |
ENSRNOT00000008840
|
Fabp1
|
fatty acid binding protein 1 |
| chr5_+_173640780 | 28.52 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr7_+_38742051 | 27.73 |
ENSRNOT00000006070
|
Dcn
|
decorin |
| chr13_-_105141030 | 27.58 |
ENSRNOT00000003313
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr4_+_30313102 | 27.54 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr1_+_47605262 | 26.64 |
ENSRNOT00000089458
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr8_-_130429132 | 26.57 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chrX_-_40086870 | 26.51 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr1_-_89488223 | 26.34 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr13_-_105140473 | 25.95 |
ENSRNOT00000087023
|
Tgfb2
|
transforming growth factor, beta 2 |
| chrX_-_23139694 | 25.46 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_+_80072489 | 25.43 |
ENSRNOT00000057176
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr12_+_45905371 | 24.64 |
ENSRNOT00000039275
|
Hspb8
|
heat shock protein family B (small) member 8 |
| chr13_-_52136127 | 23.51 |
ENSRNOT00000009398
|
Timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr4_+_64088900 | 23.28 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr1_+_18491384 | 23.24 |
ENSRNOT00000079138
ENSRNOT00000014917 |
Lama2
|
laminin subunit alpha 2 |
| chr12_+_42097626 | 22.50 |
ENSRNOT00000001893
|
Tbx5
|
T-box 5 |
| chr8_-_78233430 | 22.43 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr4_-_150829741 | 21.92 |
ENSRNOT00000051846
ENSRNOT00000052017 |
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr1_+_47605415 | 21.74 |
ENSRNOT00000034842
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr14_-_16979760 | 21.46 |
ENSRNOT00000003020
|
Stbd1
|
starch binding domain 1 |
| chr8_+_50525091 | 21.33 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr3_-_37803112 | 21.24 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr16_-_8885797 | 21.03 |
ENSRNOT00000073370
|
RGD1564899
|
similar to chromosome 10 open reading frame 71 |
| chr5_+_74766636 | 20.28 |
ENSRNOT00000030913
|
Rn50_5_0791.1
|
|
| chr2_+_27107318 | 20.14 |
ENSRNOT00000019553
|
Arhgef26
|
Rho guanine nucleotide exchange factor 26 |
| chr2_-_182038178 | 19.98 |
ENSRNOT00000040708
|
Fgb
|
fibrinogen beta chain |
| chr12_+_19196611 | 19.77 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr2_-_257474733 | 19.74 |
ENSRNOT00000016967
ENSRNOT00000066780 |
Nexn
|
nexilin (F actin binding protein) |
| chr3_+_58632476 | 19.73 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chr15_+_43298794 | 19.65 |
ENSRNOT00000012736
|
Adra1a
|
adrenoceptor alpha 1A |
| chr10_-_34439470 | 19.56 |
ENSRNOT00000072081
|
Btnl9
|
butyrophilin-like 9 |
| chr6_-_109935533 | 19.29 |
ENSRNOT00000013516
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr5_-_57372239 | 19.22 |
ENSRNOT00000012975
|
Aqp7
|
aquaporin 7 |
| chr19_-_37440471 | 19.07 |
ENSRNOT00000066636
|
Zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr2_+_61039053 | 19.05 |
ENSRNOT00000025693
|
Adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr1_+_87009730 | 18.82 |
ENSRNOT00000027537
|
Ech1
|
enoyl-CoA hydratase 1 |
| chr9_+_81968332 | 18.68 |
ENSRNOT00000023152
|
Cyp27a1
|
cytochrome P450, family 27, subfamily a, polypeptide 1 |
| chr4_-_150829913 | 18.57 |
ENSRNOT00000041571
|
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr1_-_222178725 | 18.40 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr12_-_11733099 | 18.29 |
ENSRNOT00000051244
ENSRNOT00000087257 |
Cyp3a23/3a1
|
cytochrome P450, family 3, subfamily a, polypeptide 23/polypeptide 1 |
| chr2_-_62634785 | 17.76 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr13_+_52889737 | 17.58 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr10_+_53778662 | 17.52 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr3_-_120306551 | 17.39 |
ENSRNOT00000021082
|
Mall
|
mal, T-cell differentiation protein-like |
| chr7_+_35125424 | 16.79 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr19_-_37427989 | 16.61 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr18_+_70733872 | 16.51 |
ENSRNOT00000067018
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr8_+_49441106 | 16.43 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr4_-_82203154 | 16.41 |
ENSRNOT00000086210
|
LOC100912608
|
homeobox protein Hox-A10-like |
| chr1_-_170397191 | 16.41 |
ENSRNOT00000090181
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr1_+_7252349 | 16.36 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr19_-_983556 | 16.33 |
ENSRNOT00000064781
|
Bean1
|
brain expressed, associated with NEDD4, 1 |
| chr5_-_17061837 | 16.29 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr19_+_52077109 | 16.09 |
ENSRNOT00000020225
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr8_+_71168097 | 16.01 |
ENSRNOT00000081375
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr8_-_85645718 | 15.70 |
ENSRNOT00000032185
|
Gsta2
|
glutathione S-transferase alpha 2 |
| chr1_+_248723397 | 15.43 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr18_-_48384645 | 15.15 |
ENSRNOT00000023485
|
Ppic
|
peptidylprolyl isomerase C |
| chr8_+_32018560 | 15.07 |
ENSRNOT00000007358
|
Adamts8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr3_+_63379031 | 14.86 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr19_-_37448067 | 14.83 |
ENSRNOT00000092844
|
Zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr5_+_119097715 | 14.79 |
ENSRNOT00000045987
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr4_-_123494742 | 14.66 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr6_+_23337571 | 14.51 |
ENSRNOT00000011832
|
RGD1304963
|
similar to hypothetical protein MGC38716 |
| chr17_-_55300998 | 14.45 |
ENSRNOT00000064331
|
Svil
|
supervillin |
| chr13_-_56877611 | 14.23 |
ENSRNOT00000079040
ENSRNOT00000017195 |
Cfhr1
|
complement factor H-related 1 |
| chr19_+_52077501 | 14.01 |
ENSRNOT00000079240
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr13_-_56763981 | 13.99 |
ENSRNOT00000087916
|
LOC100361907
|
complement factor H-related protein B |
| chr8_-_56393233 | 13.87 |
ENSRNOT00000016263
|
Fdx1
|
ferredoxin 1 |
| chr3_-_48536235 | 13.73 |
ENSRNOT00000083536
|
Fap
|
fibroblast activation protein, alpha |
| chr1_-_81412251 | 13.73 |
ENSRNOT00000026946
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr9_+_97355924 | 13.70 |
ENSRNOT00000026558
|
Ackr3
|
atypical chemokine receptor 3 |
| chr12_-_30566032 | 13.32 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr1_-_90520012 | 13.27 |
ENSRNOT00000028698
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr12_-_5822874 | 13.27 |
ENSRNOT00000075920
|
Fry
|
FRY microtubule binding protein |
| chr1_-_241155537 | 13.19 |
ENSRNOT00000034216
ENSRNOT00000073493 |
Mamdc2
|
MAM domain containing 2 |
| chr18_+_38292701 | 13.16 |
ENSRNOT00000037796
|
Scgb3a2
|
secretoglobin, family 3A, member 2 |
| chr5_-_100647727 | 13.09 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr20_+_28989491 | 13.05 |
ENSRNOT00000074524
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr6_+_56846789 | 13.03 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr9_-_19613360 | 12.96 |
ENSRNOT00000029593
|
Rcan2
|
regulator of calcineurin 2 |
| chr10_+_37582706 | 12.93 |
ENSRNOT00000007630
|
RGD1306484
|
similar to growth and transformation-dependent protein |
| chr7_+_11490852 | 12.92 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr17_+_12261102 | 12.81 |
ENSRNOT00000015525
|
Nfil3
|
nuclear factor, interleukin 3 regulated |
| chr7_-_29701586 | 12.74 |
ENSRNOT00000009084
ENSRNOT00000089269 |
Ano4
|
anoctamin 4 |
| chr8_+_71167305 | 12.62 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr5_+_159484370 | 12.62 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr9_+_17340341 | 12.45 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chr13_-_47377703 | 12.43 |
ENSRNOT00000005461
|
C4bpa
|
complement component 4 binding protein, alpha |
| chrX_+_120624518 | 12.41 |
ENSRNOT00000007967
|
Slc6a14
|
solute carrier family 6 member 14 |
| chr6_-_41870046 | 12.37 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr8_-_119889661 | 12.32 |
ENSRNOT00000011780
|
Stac
|
SH3 and cysteine rich domain |
| chr15_+_10120206 | 12.32 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chrX_-_142248369 | 12.28 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr5_-_100647298 | 12.26 |
ENSRNOT00000067538
ENSRNOT00000013092 |
Nfib
|
nuclear factor I/B |
| chr1_-_127337882 | 12.24 |
ENSRNOT00000085158
|
Aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr13_-_89518939 | 12.20 |
ENSRNOT00000004228
|
Sdhc
|
succinate dehydrogenase complex subunit C |
| chr20_-_4489281 | 12.19 |
ENSRNOT00000031548
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr2_-_231521052 | 12.11 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr2_-_196113149 | 12.04 |
ENSRNOT00000088465
|
Selenbp1
|
selenium binding protein 1 |
| chr7_+_144647587 | 11.97 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr4_+_68653369 | 11.95 |
ENSRNOT00000046917
|
Tas2r137
|
taste receptor, type 2, member 137 |
| chr1_+_31264755 | 11.91 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr19_-_57699113 | 11.91 |
ENSRNOT00000026767
|
Egln1
|
egl-9 family hypoxia-inducible factor 1 |
| chr4_-_100783750 | 11.79 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr17_+_22620721 | 11.55 |
ENSRNOT00000019478
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr9_-_9702306 | 11.52 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr9_+_100511610 | 11.44 |
ENSRNOT00000033233
|
Ano7
|
anoctamin 7 |
| chr7_+_34533543 | 11.18 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr10_-_89187474 | 11.12 |
ENSRNOT00000064931
|
Usmg5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr7_+_76059386 | 10.98 |
ENSRNOT00000009337
|
Grhl2
|
grainyhead-like transcription factor 2 |
| chrX_-_72515320 | 10.98 |
ENSRNOT00000076725
|
Phka1
|
phosphorylase kinase, alpha 1 |
| chr3_+_151126591 | 10.97 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr3_+_79678201 | 10.89 |
ENSRNOT00000087604
ENSRNOT00000079709 |
Mtch2
|
mitochondrial carrier 2 |
| chrX_+_139780386 | 10.82 |
ENSRNOT00000081147
|
AABR07041779.2
|
|
| chr19_-_22281778 | 10.74 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_-_90520344 | 10.68 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr13_+_48575091 | 10.65 |
ENSRNOT00000009838
|
Pm20d1
|
peptidase M20 domain containing 1 |
| chr16_+_16949232 | 10.56 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr2_-_243407608 | 10.52 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr3_+_122544788 | 10.47 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr3_+_54253949 | 10.39 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr9_-_9985630 | 10.37 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr6_-_105097054 | 10.34 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr2_-_14701903 | 10.32 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr10_-_38782419 | 10.32 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr10_-_102289837 | 10.30 |
ENSRNOT00000044922
|
AABR07030729.1
|
|
| chr3_+_132052612 | 10.30 |
ENSRNOT00000030148
|
AABR07053879.1
|
|
| chrX_-_139464798 | 10.20 |
ENSRNOT00000003282
|
Gpc4
|
glypican 4 |
| chr17_+_49417067 | 10.19 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr7_+_74047814 | 10.17 |
ENSRNOT00000014879
|
Osr2
|
odd-skipped related transciption factor 2 |
| chr8_-_55087832 | 10.11 |
ENSRNOT00000032152
|
Dlat
|
dihydrolipoamide S-acetyltransferase |
| chr8_-_94352246 | 10.09 |
ENSRNOT00000013244
|
Me1
|
malic enzyme 1 |
| chr7_-_117576737 | 10.00 |
ENSRNOT00000039795
|
Dgat1
|
diacylglycerol O-acyltransferase 1 |
| chr1_+_193537137 | 9.87 |
ENSRNOT00000029967
|
AABR07005667.1
|
|
| chr2_-_178389608 | 9.85 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr1_-_167911961 | 9.82 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr1_+_197839430 | 9.80 |
ENSRNOT00000025043
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr19_-_25134055 | 9.79 |
ENSRNOT00000007519
|
Misp3
|
MISP family member 3 |
| chr7_-_74735650 | 9.77 |
ENSRNOT00000014407
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr1_-_219144610 | 9.67 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr1_+_282557426 | 9.62 |
ENSRNOT00000088966
|
AABR07007134.1
|
|
| chr11_+_70687500 | 9.61 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr8_-_116993193 | 9.57 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr8_-_49227273 | 9.54 |
ENSRNOT00000050878
|
Atp5l
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr11_+_61083757 | 9.52 |
ENSRNOT00000002790
|
Boc
|
BOC cell adhesion associated, oncogene regulated |
| chr1_+_72420352 | 9.43 |
ENSRNOT00000066307
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr8_+_59507990 | 9.38 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chrX_-_72132886 | 9.24 |
ENSRNOT00000077678
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chrX_-_72132495 | 9.20 |
ENSRNOT00000076391
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr11_-_79703736 | 9.13 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr13_+_56598957 | 9.09 |
ENSRNOT00000016944
ENSRNOT00000080335 ENSRNOT00000089913 |
F13b
|
coagulation factor XIII B chain |
| chr18_+_27956429 | 9.02 |
ENSRNOT00000080999
|
Ctnna1
|
catenin alpha 1 |
| chr8_+_117679278 | 9.01 |
ENSRNOT00000042114
|
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein I |
| chr4_+_61771970 | 8.95 |
ENSRNOT00000078996
ENSRNOT00000013073 |
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr4_+_14039977 | 8.86 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr3_-_12007570 | 8.75 |
ENSRNOT00000060186
|
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr15_-_39886613 | 8.66 |
ENSRNOT00000089963
|
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr7_-_124041594 | 8.66 |
ENSRNOT00000064700
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr1_-_89017290 | 8.62 |
ENSRNOT00000028438
|
Psenen
|
presenilin enhancer gamma secretase subunit |
| chr2_+_60337667 | 8.61 |
ENSRNOT00000024035
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr5_+_134492756 | 8.59 |
ENSRNOT00000012888
ENSRNOT00000057095 ENSRNOT00000051385 |
Cyp4a1
|
cytochrome P450, family 4, subfamily a, polypeptide 1 |
| chr15_+_19603288 | 8.57 |
ENSRNOT00000035491
|
LOC102552640
|
REST corepressor 2-like |
| chr6_+_73358112 | 8.49 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr3_+_12262822 | 8.48 |
ENSRNOT00000022585
|
Angptl2
|
angiopoietin-like 2 |
| chr13_-_103080920 | 8.46 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr5_-_101138427 | 8.46 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr2_+_158097843 | 8.43 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr10_-_36419926 | 8.39 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr9_-_55256340 | 8.32 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr4_+_102426224 | 8.25 |
ENSRNOT00000073768
|
LOC103694107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 pseudogene |
| chr18_+_61261418 | 8.18 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr11_+_57430166 | 8.17 |
ENSRNOT00000093201
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr4_+_122244711 | 8.13 |
ENSRNOT00000038251
|
Uroc1
|
urocanate hydratase 1 |
| chr6_-_7961207 | 8.04 |
ENSRNOT00000007174
|
Abcg5
|
ATP binding cassette subfamily G member 5 |
| chr11_-_62451149 | 8.02 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_-_42127678 | 7.98 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr18_+_64114933 | 7.96 |
ENSRNOT00000022364
|
Mc5r
|
melanocortin 5 receptor |
| chr8_-_47339343 | 7.91 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr8_-_22542997 | 7.87 |
ENSRNOT00000061009
|
Tmed1
|
transmembrane p24 trafficking protein 1 |
| chr5_+_124476168 | 7.82 |
ENSRNOT00000077754
|
RGD1564074
|
similar to novel protein |
| chr1_+_282715344 | 7.79 |
ENSRNOT00000074399
|
LOC100910259
|
liver carboxylesterase-like |
| chr10_+_56662561 | 7.78 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr2_-_119140110 | 7.76 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1i2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.8 | 53.5 | GO:0070237 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 10.6 | 31.8 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 10.5 | 41.9 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 10.3 | 61.6 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 10.1 | 40.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 9.6 | 28.8 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 7.1 | 21.3 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 6.7 | 40.5 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 6.6 | 19.7 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 6.5 | 19.6 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 6.4 | 19.3 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 6.3 | 25.4 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 6.2 | 18.7 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 5.8 | 46.8 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 5.4 | 16.3 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 5.3 | 26.6 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 5.3 | 26.3 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 4.8 | 24.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 4.7 | 23.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 4.6 | 18.4 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 4.6 | 23.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 4.6 | 13.7 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 4.5 | 22.5 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 4.5 | 13.5 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 4.5 | 9.0 | GO:0016487 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 4.1 | 12.4 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of lymphangiogenesis(GO:1901492) |
| 4.1 | 12.4 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 4.1 | 16.5 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 4.1 | 12.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 4.0 | 20.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 4.0 | 12.0 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 4.0 | 8.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 3.8 | 19.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 3.6 | 29.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 3.6 | 10.7 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 3.4 | 10.3 | GO:0021539 | subthalamus development(GO:0021539) |
| 3.3 | 10.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 3.3 | 13.3 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 3.3 | 9.8 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 3.2 | 25.5 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 3.0 | 12.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 3.0 | 9.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 3.0 | 29.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 3.0 | 11.9 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 2.9 | 14.4 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 2.9 | 8.6 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) |
| 2.7 | 11.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 2.7 | 48.4 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 2.7 | 39.8 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 2.6 | 5.2 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 2.4 | 19.4 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 2.4 | 4.8 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 2.3 | 16.4 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 2.3 | 6.8 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 2.3 | 6.8 | GO:0043132 | phospholipid transfer to membrane(GO:0006649) NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) |
| 2.2 | 28.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 2.2 | 6.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 2.2 | 19.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 2.2 | 8.8 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 2.1 | 27.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 2.1 | 8.4 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 2.1 | 6.3 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 2.1 | 12.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 2.0 | 15.8 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.9 | 17.4 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.9 | 7.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 1.9 | 7.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 1.9 | 11.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.8 | 20.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 1.8 | 14.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 1.8 | 23.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 1.8 | 16.0 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.8 | 12.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 1.7 | 17.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.7 | 6.8 | GO:1990375 | baculum development(GO:1990375) |
| 1.7 | 11.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.7 | 3.4 | GO:0061354 | cardiac right atrium morphogenesis(GO:0003213) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 1.7 | 39.9 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 1.6 | 6.6 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 1.6 | 27.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.6 | 8.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 1.6 | 27.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 1.6 | 10.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 1.5 | 12.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 1.5 | 6.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 1.5 | 23.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 1.5 | 17.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.4 | 4.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 1.4 | 10.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 1.4 | 7.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.4 | 4.3 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.4 | 5.7 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 1.4 | 18.3 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 1.4 | 4.2 | GO:1901295 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 1.3 | 5.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.3 | 3.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.3 | 5.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 1.3 | 5.0 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 1.2 | 3.7 | GO:0042335 | cuticle development(GO:0042335) |
| 1.2 | 15.7 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 1.2 | 10.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 1.2 | 4.6 | GO:0048252 | lauric acid metabolic process(GO:0048252) |
| 1.1 | 28.5 | GO:0014850 | response to muscle activity(GO:0014850) |
| 1.1 | 34.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 1.1 | 15.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.1 | 6.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 1.1 | 13.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.1 | 12.9 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 1.1 | 10.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 1.0 | 29.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 1.0 | 10.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 1.0 | 4.0 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 1.0 | 6.0 | GO:0061525 | hindgut development(GO:0061525) |
| 1.0 | 13.0 | GO:0007614 | short-term memory(GO:0007614) |
| 1.0 | 21.5 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 1.0 | 5.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.0 | 5.8 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 1.0 | 7.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.9 | 12.2 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.9 | 13.8 | GO:1901201 | regulation of extracellular matrix assembly(GO:1901201) |
| 0.9 | 29.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.9 | 9.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.9 | 1.8 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.9 | 2.6 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.8 | 5.9 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.8 | 9.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.8 | 4.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.8 | 34.7 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.7 | 10.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.7 | 2.2 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.7 | 2.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.7 | 3.5 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.7 | 6.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 18.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.6 | 3.0 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.6 | 11.5 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.6 | 6.2 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.6 | 5.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.6 | 6.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.5 | 15.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.5 | 14.8 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.5 | 12.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.5 | 6.9 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.5 | 10.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.5 | 4.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.5 | 9.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.5 | 5.0 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.5 | 3.5 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.5 | 2.0 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 1.5 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.5 | 1.4 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.5 | 1.8 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 6.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.4 | 13.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.4 | 13.1 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.4 | 23.1 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.4 | 4.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 4.7 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.4 | 6.6 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.4 | 9.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.4 | 1.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.4 | 2.2 | GO:0009820 | alkaloid metabolic process(GO:0009820) quinolinate metabolic process(GO:0046874) |
| 0.4 | 0.7 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.4 | 18.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 2.8 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.3 | 2.8 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.3 | 10.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.3 | 1.2 | GO:1902861 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.3 | 8.5 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.3 | 1.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.3 | 2.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 0.6 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.3 | 15.0 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.3 | 0.5 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 1.9 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.3 | 8.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 16.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 12.9 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.2 | 8.1 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.2 | 8.0 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.2 | 2.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 3.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 1.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 2.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 0.2 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.2 | 1.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 3.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 4.0 | GO:0003416 | endochondral bone growth(GO:0003416) bone growth(GO:0098868) |
| 0.2 | 7.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 6.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 3.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 19.3 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.2 | 4.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 0.5 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 1.4 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.2 | 3.0 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 2.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 1.2 | GO:0006729 | L-phenylalanine catabolic process(GO:0006559) tetrahydrobiopterin biosynthetic process(GO:0006729) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 3.5 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.1 | 12.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 3.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 3.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.1 | 3.9 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 6.7 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 4.8 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 3.8 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 4.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 4.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.4 | GO:0072003 | kidney rudiment formation(GO:0072003) metanephric tubule formation(GO:0072174) |
| 0.1 | 1.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.3 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.9 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.1 | 4.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 114.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 3.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.2 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 2.6 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.0 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.1 | 3.4 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 5.0 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 3.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.7 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 2.4 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.1 | 0.7 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 2.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 4.5 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 13.2 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.1 | 0.8 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 0.2 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 1.0 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 2.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 18.0 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 5.1 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 1.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 5.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 3.1 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.8 | 59.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 9.9 | 39.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 6.2 | 24.8 | GO:0045281 | respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) |
| 5.8 | 34.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 4.7 | 23.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 4.1 | 16.4 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 3.9 | 19.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 3.4 | 27.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 3.4 | 34.4 | GO:0045179 | apical cortex(GO:0045179) |
| 3.4 | 40.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 2.7 | 21.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 2.7 | 8.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 2.5 | 25.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 2.5 | 20.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.5 | 12.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 2.3 | 23.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 2.3 | 97.8 | GO:0043034 | costamere(GO:0043034) |
| 1.9 | 17.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.9 | 40.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 1.8 | 11.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.8 | 19.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 1.7 | 5.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 1.5 | 9.3 | GO:0030478 | actin cap(GO:0030478) |
| 1.5 | 16.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.5 | 19.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 1.4 | 8.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 1.3 | 27.7 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 1.3 | 6.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.2 | 15.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.1 | 6.8 | GO:0032059 | bleb(GO:0032059) |
| 1.1 | 10.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 1.1 | 109.5 | GO:0005604 | basement membrane(GO:0005604) |
| 1.1 | 75.7 | GO:0030315 | T-tubule(GO:0030315) |
| 1.0 | 20.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.0 | 6.2 | GO:0089701 | U2AF(GO:0089701) |
| 1.0 | 3.9 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.9 | 9.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.9 | 3.4 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.8 | 17.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.8 | 13.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.8 | 41.7 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.7 | 42.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.7 | 4.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.7 | 42.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.6 | 22.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.6 | 2.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.6 | 3.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.6 | 2.9 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.6 | 15.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 5.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.6 | 10.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.5 | 3.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.5 | 4.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.5 | 80.4 | GO:0030016 | myofibril(GO:0030016) |
| 0.5 | 8.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 2.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.5 | 51.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.5 | 34.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.5 | 2.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.5 | 8.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 9.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 19.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.4 | 9.9 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 27.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 16.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 2.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.3 | 12.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.3 | 6.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.3 | 3.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 2.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 4.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.2 | 2.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 9.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 2.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 37.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 11.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 66.1 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.2 | 1.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 2.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 22.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 3.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 11.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 7.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 40.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 3.1 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 19.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 23.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 4.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 4.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 3.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 32.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 6.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 7.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 7.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 4.1 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 17.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 72.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 9.9 | 39.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 8.0 | 31.8 | GO:0035877 | death effector domain binding(GO:0035877) |
| 7.8 | 23.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 7.2 | 21.5 | GO:2001070 | starch binding(GO:2001070) |
| 7.1 | 21.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 6.7 | 40.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 6.5 | 19.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 6.4 | 19.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 6.3 | 31.6 | GO:0019862 | IgA binding(GO:0019862) |
| 6.2 | 24.8 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 5.3 | 31.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 5.1 | 5.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 3.6 | 25.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 3.4 | 10.1 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 3.2 | 13.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 3.0 | 6.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 3.0 | 9.0 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 2.7 | 16.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 2.7 | 21.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 2.6 | 23.5 | GO:0015450 | protein channel activity(GO:0015266) P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 2.6 | 13.0 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 2.6 | 51.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 2.5 | 10.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 2.5 | 22.5 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 2.5 | 24.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 2.5 | 9.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 2.4 | 16.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 2.2 | 41.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 2.1 | 49.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 2.1 | 10.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 2.1 | 16.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 2.0 | 12.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 2.0 | 16.1 | GO:0008430 | selenium binding(GO:0008430) |
| 1.9 | 5.8 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 1.9 | 29.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.9 | 7.5 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 1.9 | 13.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 1.8 | 18.4 | GO:0050693 | LBD domain binding(GO:0050693) |
| 1.8 | 5.5 | GO:0047708 | biotinidase activity(GO:0047708) |
| 1.8 | 8.9 | GO:0070538 | oleic acid binding(GO:0070538) lipoteichoic acid receptor activity(GO:0070892) |
| 1.7 | 8.6 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 1.7 | 5.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 1.6 | 6.6 | GO:0055100 | adiponectin binding(GO:0055100) |
| 1.6 | 6.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 1.5 | 11.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 1.5 | 10.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.4 | 17.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.4 | 8.7 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.4 | 27.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 1.4 | 12.4 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 1.4 | 6.8 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.3 | 10.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.3 | 24.5 | GO:0001848 | complement binding(GO:0001848) |
| 1.3 | 9.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 1.3 | 6.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.3 | 7.6 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 1.3 | 10.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 1.3 | 6.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.2 | 8.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 1.2 | 17.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.2 | 3.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.1 | 5.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.1 | 12.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 1.1 | 6.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.1 | 8.8 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 1.0 | 10.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 1.0 | 11.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 1.0 | 3.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.0 | 26.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.0 | 3.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.9 | 3.8 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.9 | 7.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.9 | 29.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.9 | 33.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.9 | 15.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.9 | 12.0 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.9 | 3.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.8 | 6.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.8 | 15.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.8 | 6.8 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.7 | 25.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.7 | 2.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.7 | 2.2 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.7 | 4.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.7 | 1.4 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.7 | 8.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.7 | 5.4 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.7 | 11.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.7 | 3.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.6 | 3.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.6 | 5.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 5.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.6 | 6.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.6 | 6.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 32.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.6 | 14.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.6 | 4.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.6 | 10.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.6 | 1.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 1.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.6 | 4.0 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.6 | 6.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.6 | 16.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 6.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.5 | 20.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 39.9 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.5 | 10.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 6.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 2.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 19.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.5 | 27.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.5 | 2.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.5 | 3.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.5 | 1.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.4 | 3.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.4 | 1.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 11.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.4 | 1.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.4 | 30.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 2.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 48.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 3.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 7.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 5.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 9.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 4.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 1.8 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.3 | 2.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 0.6 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.3 | 3.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 4.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.3 | 6.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 20.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 8.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 13.7 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.2 | 1.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 10.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 2.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 3.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 4.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.8 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.2 | 3.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 4.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 8.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.2 | 2.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 8.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 5.0 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 13.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 1.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 6.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 3.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 3.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 17.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 12.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 13.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 7.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 1.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 44.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.7 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 2.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 68.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 1.0 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.1 | 4.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.2 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.1 | 2.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 2.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 12.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 7.4 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 3.3 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 7.5 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 2.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 4.9 | GO:0016491 | oxidoreductase activity(GO:0016491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 53.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 2.1 | 6.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 1.6 | 17.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 1.4 | 23.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.4 | 9.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.7 | 27.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.7 | 41.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.6 | 27.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.6 | 32.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.6 | 11.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.6 | 13.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.6 | 25.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.6 | 17.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.6 | 11.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.5 | 9.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.5 | 9.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.4 | 11.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.4 | 6.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 12.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.4 | 3.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.4 | 6.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 4.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.4 | 8.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 10.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 15.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 11.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 65.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 8.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 9.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 8.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 12.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 38.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 8.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 6.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 8.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 6.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 6.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 34.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.0 | 27.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.8 | 29.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.6 | 19.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.6 | 61.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.6 | 18.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.5 | 17.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.4 | 5.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.4 | 91.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 1.2 | 19.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 1.2 | 46.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 1.1 | 25.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 1.1 | 21.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.0 | 6.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.0 | 12.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.9 | 81.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.9 | 12.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.9 | 13.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.9 | 8.6 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.8 | 23.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.7 | 36.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.7 | 10.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.7 | 10.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.7 | 8.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 6.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 9.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.6 | 11.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.6 | 20.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.6 | 23.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.5 | 4.3 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.5 | 11.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 57.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.5 | 17.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 8.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 10.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 5.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.4 | 12.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 5.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 11.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 7.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 16.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 17.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 6.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 2.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 14.7 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.2 | 3.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 4.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 3.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 2.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 2.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 4.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.2 | 19.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 22.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 10.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 3.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |