Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
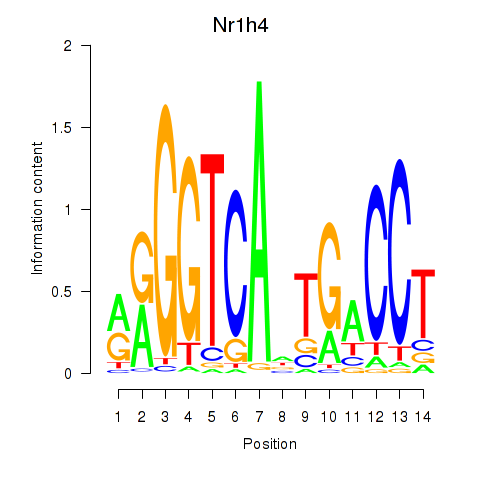
Results for Nr1h4
Z-value: 1.56
Transcription factors associated with Nr1h4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1h4
|
ENSRNOG00000007197 | nuclear receptor subfamily 1, group H, member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h4 | rn6_v1_chr7_-_30105132_30105132 | 0.60 | 4.7e-33 | Click! |
Activity profile of Nr1h4 motif
Sorted Z-values of Nr1h4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_55587946 | 182.67 |
ENSRNOT00000075107
|
Abcb11
|
ATP binding cassette subfamily B member 11 |
| chr16_-_7007287 | 146.79 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr16_-_7007051 | 143.69 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr9_+_74124016 | 142.46 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr7_+_2689501 | 123.29 |
ENSRNOT00000041341
|
Apof
|
apolipoprotein F |
| chr1_-_170431073 | 117.67 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr13_+_89597138 | 91.99 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr7_-_123655896 | 91.06 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr2_+_248649441 | 63.57 |
ENSRNOT00000067165
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr9_-_4945352 | 62.39 |
ENSRNOT00000082530
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr20_-_4542073 | 61.17 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr8_+_116857684 | 54.33 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr2_+_20857202 | 54.31 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr1_+_100470722 | 53.41 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr6_+_107550904 | 51.20 |
ENSRNOT00000013760
|
Acot5
|
acyl-CoA thioesterase 5 |
| chr1_-_80617057 | 50.78 |
ENSRNOT00000080453
|
Apoe
|
apolipoprotein E |
| chr4_+_61850348 | 44.97 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr4_+_175814118 | 42.58 |
ENSRNOT00000013409
ENSRNOT00000013514 |
Slco1b2
|
solute carrier organic anion transporter family, member 1B2 |
| chrX_+_6430594 | 40.08 |
ENSRNOT00000044009
|
Maob
|
monoamine oxidase B |
| chr10_-_104748003 | 40.05 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr17_-_42127678 | 38.81 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr1_-_89488223 | 37.84 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr11_+_83868655 | 35.16 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr5_+_151776004 | 34.97 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr7_-_102298522 | 34.20 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr3_-_121835360 | 34.13 |
ENSRNOT00000006126
ENSRNOT00000080818 |
Il1a
|
interleukin 1 alpha |
| chr6_-_10899200 | 33.57 |
ENSRNOT00000089104
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr8_+_115151627 | 31.01 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr7_-_124999137 | 27.64 |
ENSRNOT00000039228
|
Pnpla5
|
patatin-like phospholipase domain containing 5 |
| chr4_+_100277391 | 23.94 |
ENSRNOT00000086820
ENSRNOT00000084281 ENSRNOT00000017927 |
Ggcx
|
gamma-glutamyl carboxylase |
| chr1_+_264741911 | 22.79 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr1_-_98486976 | 21.82 |
ENSRNOT00000024083
|
Etfb
|
electron transfer flavoprotein beta subunit |
| chr10_-_46582854 | 20.81 |
ENSRNOT00000004753
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr16_-_6404957 | 19.18 |
ENSRNOT00000048459
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr1_+_219833299 | 18.35 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chrX_-_139464798 | 18.33 |
ENSRNOT00000003282
|
Gpc4
|
glypican 4 |
| chr5_+_136683592 | 17.41 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chrX_+_21696772 | 17.26 |
ENSRNOT00000043559
|
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr9_+_99795678 | 15.93 |
ENSRNOT00000056601
|
Olr1353
|
olfactory receptor 1353 |
| chr16_-_6404578 | 15.56 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr3_-_39596718 | 14.73 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr11_-_31772984 | 13.63 |
ENSRNOT00000002771
|
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr7_-_12882753 | 13.14 |
ENSRNOT00000011275
ENSRNOT00000044275 |
Bsg
|
basigin |
| chr9_+_61810859 | 12.97 |
ENSRNOT00000079058
|
AABR07067763.1
|
|
| chr12_+_9034308 | 11.63 |
ENSRNOT00000001248
|
Flt1
|
FMS-related tyrosine kinase 1 |
| chr18_+_16616937 | 11.63 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chr1_+_72420352 | 11.23 |
ENSRNOT00000066307
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr7_-_118474609 | 11.12 |
ENSRNOT00000089076
|
AABR07058464.1
|
|
| chr8_+_117455262 | 10.96 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr1_-_198794947 | 10.64 |
ENSRNOT00000024639
|
Znf768
|
zinc finger protein 768 |
| chr14_+_36216002 | 10.37 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr1_+_255479261 | 9.79 |
ENSRNOT00000079808
|
Tnks2
|
tankyrase 2 |
| chr10_+_37582706 | 9.46 |
ENSRNOT00000007630
|
RGD1306484
|
similar to growth and transformation-dependent protein |
| chr3_+_8958090 | 8.97 |
ENSRNOT00000086789
ENSRNOT00000064557 |
Dolpp1
|
dolichyldiphosphatase 1 |
| chr13_-_89619398 | 8.95 |
ENSRNOT00000058423
|
Ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr14_+_2325308 | 8.87 |
ENSRNOT00000000072
|
Atp5i
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr10_-_109333899 | 8.79 |
ENSRNOT00000006225
|
Slc38a10
|
solute carrier family 38, member 10 |
| chr3_-_77961992 | 8.72 |
ENSRNOT00000089105
|
AC132028.2
|
|
| chr3_+_77921705 | 8.70 |
ENSRNOT00000064006
|
AC132028.1
|
|
| chr14_-_33580566 | 8.39 |
ENSRNOT00000063942
|
Paics
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr3_+_77584990 | 8.26 |
ENSRNOT00000046926
|
Olr664
|
olfactory receptor 664 |
| chr18_+_57011575 | 8.08 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr3_-_77696294 | 8.01 |
ENSRNOT00000050849
|
Olr670
|
olfactory receptor 670 |
| chr1_-_219259448 | 7.67 |
ENSRNOT00000024517
|
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr8_-_128622314 | 7.29 |
ENSRNOT00000084950
ENSRNOT00000024408 |
Gorasp1
|
golgi reassembly stacking protein 1 |
| chr18_-_62965538 | 7.20 |
ENSRNOT00000025164
|
Mppe1
|
metallophosphoesterase 1 |
| chr9_+_64095978 | 7.14 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr19_-_15733412 | 6.93 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr6_+_107245820 | 6.62 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr5_-_154319629 | 6.55 |
ENSRNOT00000014415
|
Lypla2
|
lysophospholipase II |
| chr1_-_22548675 | 6.39 |
ENSRNOT00000061209
|
Taar5
|
trace amine-associated receptor 5 |
| chr3_-_77883067 | 6.27 |
ENSRNOT00000087979
|
Olr677
|
olfactory receptor 677 |
| chr12_-_22726982 | 6.25 |
ENSRNOT00000001921
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr4_+_68656928 | 6.17 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr1_-_266934303 | 6.02 |
ENSRNOT00000036202
|
Calhm3
|
calcium homeostasis modulator 3 |
| chr15_+_61662264 | 5.86 |
ENSRNOT00000072969
|
Mtrf1
|
mitochondrial translation release factor 1 |
| chr6_+_99625306 | 5.50 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr4_+_7314369 | 5.06 |
ENSRNOT00000029724
|
Atg9b
|
autophagy related 9B |
| chr10_+_56576428 | 5.03 |
ENSRNOT00000079237
ENSRNOT00000023291 |
Cldn7
|
claudin 7 |
| chr3_-_77605962 | 4.92 |
ENSRNOT00000090062
|
Olr665
|
olfactory receptor 665 |
| chr8_+_23014956 | 4.90 |
ENSRNOT00000018009
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr19_-_41567013 | 4.85 |
ENSRNOT00000022757
|
Zfp612
|
zinc finger protein 612 |
| chr1_-_64099277 | 4.82 |
ENSRNOT00000084846
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr14_+_33580541 | 4.75 |
ENSRNOT00000002905
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr6_+_124123228 | 4.73 |
ENSRNOT00000005329
|
Psmc1
|
proteasome 26S subunit, ATPase 1 |
| chr14_+_48764670 | 4.52 |
ENSRNOT00000090271
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_-_60864478 | 4.51 |
ENSRNOT00000088653
|
AABR07038795.1
|
|
| chr3_-_37947283 | 4.46 |
ENSRNOT00000009181
|
Arl5a
|
ADP-ribosylation factor like GTPase 5A |
| chr9_+_10004805 | 4.45 |
ENSRNOT00000074117
|
Slc25a41
|
solute carrier family 25, member 41 |
| chr10_-_109913879 | 4.32 |
ENSRNOT00000072947
|
LOC688320
|
similar to Lung carbonyl reductase [NADPH] (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27) |
| chr2_+_240668195 | 4.28 |
ENSRNOT00000087371
|
Manba
|
mannosidase beta |
| chr3_-_78066123 | 4.24 |
ENSRNOT00000073165
|
Olr690
|
olfactory receptor 690 |
| chr7_-_117326551 | 3.92 |
ENSRNOT00000086087
|
AC107096.1
|
|
| chr1_+_100447998 | 3.82 |
ENSRNOT00000026259
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr14_-_83776863 | 3.49 |
ENSRNOT00000026481
|
Smtn
|
smoothelin |
| chrM_+_8599 | 3.22 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr10_-_29196252 | 3.20 |
ENSRNOT00000083910
|
Fabp6
|
fatty acid binding protein 6 |
| chr3_-_151371501 | 3.13 |
ENSRNOT00000073178
|
Fam83c
|
family with sequence similarity 83, member C |
| chr20_+_11114164 | 2.98 |
ENSRNOT00000001602
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr3_-_79165698 | 2.86 |
ENSRNOT00000032617
|
Olr744
|
olfactory receptor 744 |
| chr12_+_47024442 | 2.75 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr9_+_43830630 | 2.66 |
ENSRNOT00000084742
|
Cnga3
|
cyclic nucleotide gated channel alpha 3 |
| chr18_-_63185510 | 2.56 |
ENSRNOT00000024632
|
Afg3l2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chrX_+_44830849 | 2.34 |
ENSRNOT00000079680
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr14_+_88549947 | 2.10 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr7_+_54031316 | 2.02 |
ENSRNOT00000039096
|
Bbs10
|
Bardet-Biedl syndrome 10 |
| chr7_+_130542202 | 1.85 |
ENSRNOT00000079501
ENSRNOT00000045647 |
Acr
|
acrosin |
| chr3_-_153468221 | 1.73 |
ENSRNOT00000010443
|
Ghrh
|
growth hormone releasing hormone |
| chr5_+_59228199 | 1.64 |
ENSRNOT00000060282
|
Hrct1
|
histidine rich carboxyl terminus 1 |
| chr7_-_12609868 | 1.61 |
ENSRNOT00000016396
|
Kiss1r
|
KISS1 receptor |
| chr1_-_227047290 | 1.48 |
ENSRNOT00000037226
|
Ms4a15
|
membrane spanning 4-domains A15 |
| chr9_+_82647071 | 1.41 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr8_-_25829569 | 1.32 |
ENSRNOT00000071884
|
Dpy19l2
|
dpy-19 like 2 |
| chr10_+_11240138 | 1.32 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr17_-_1093873 | 1.29 |
ENSRNOT00000086130
|
Ptch1
|
patched 1 |
| chr18_-_36579403 | 1.08 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr15_-_25521393 | 1.08 |
ENSRNOT00000085910
|
Otx2
|
orthodenticle homeobox 2 |
| chr7_-_30344426 | 1.05 |
ENSRNOT00000002638
ENSRNOT00000082016 |
Scyl2
|
SCY1 like pseudokinase 2 |
| chr5_+_36024649 | 1.03 |
ENSRNOT00000013384
|
Coq3
|
coenzyme Q3 methyltransferase |
| chr3_-_77677349 | 0.99 |
ENSRNOT00000084989
|
Olr669
|
olfactory receptor 669 |
| chr10_+_61033113 | 0.93 |
ENSRNOT00000043016
|
Olr1513
|
olfactory receptor 1513 |
| chr20_+_5441876 | 0.79 |
ENSRNOT00000092476
|
Rps18
|
ribosomal protein S18 |
| chr13_-_84793463 | 0.54 |
ENSRNOT00000076886
|
Fmo9
|
flavin containing monooxygenase 9 |
| chr15_+_27207645 | 0.54 |
ENSRNOT00000011549
|
Olr1614
|
olfactory receptor 1614 |
| chr12_+_22727335 | 0.47 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr5_-_5691794 | 0.46 |
ENSRNOT00000089152
|
AABR07046763.1
|
|
| chr7_-_12609694 | 0.46 |
ENSRNOT00000093287
ENSRNOT00000079485 |
Kiss1r
|
KISS1 receptor |
| chr13_-_68858781 | 0.31 |
ENSRNOT00000065944
ENSRNOT00000092053 |
Rnf2
|
ring finger protein 2 |
| chr5_-_164585634 | 0.28 |
ENSRNOT00000065985
|
Mfn2
|
mitofusin 2 |
| chr1_-_170628915 | 0.26 |
ENSRNOT00000042865
|
Dchs1
|
dachsous cadherin-related 1 |
| chr9_+_61655963 | 0.16 |
ENSRNOT00000040461
|
Coq10b
|
coenzyme Q10B |
| chr3_+_76468294 | 0.08 |
ENSRNOT00000037779
|
Olr619
|
olfactory receptor 619 |
| chr20_+_11436267 | 0.06 |
ENSRNOT00000001631
|
Trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1h4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 30.7 | 92.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 29.4 | 117.7 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 28.5 | 142.5 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 26.1 | 182.7 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 19.4 | 38.8 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 17.8 | 53.4 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 17.1 | 51.2 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 16.9 | 50.8 | GO:1903000 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 13.3 | 40.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 12.7 | 63.6 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 12.6 | 37.8 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 11.7 | 35.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 11.4 | 34.1 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 8.8 | 290.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 6.0 | 54.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 6.0 | 23.9 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 5.6 | 61.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 5.0 | 40.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 4.6 | 18.3 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 3.9 | 42.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 3.9 | 34.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 2.9 | 11.6 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 2.7 | 11.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 2.7 | 124.6 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 2.6 | 20.8 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 2.4 | 7.1 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 2.2 | 13.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 2.1 | 6.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 2.0 | 21.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.6 | 6.5 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 1.6 | 17.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 1.5 | 54.3 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 1.4 | 91.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 1.3 | 5.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 1.2 | 4.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.1 | 27.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 1.1 | 11.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.8 | 17.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.6 | 6.9 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.6 | 8.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.6 | 13.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.6 | 13.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.5 | 1.1 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.5 | 1.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.5 | 4.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.4 | 2.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.4 | 35.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.4 | 1.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.4 | 5.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 2.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.3 | 1.3 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 1.3 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 | 5.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.3 | 4.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 8.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 1.7 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.3 | 15.4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.3 | 7.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.3 | 1.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.3 | 4.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.2 | 2.7 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.2 | 6.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 7.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 48.8 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.2 | 11.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.3 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 0.5 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 8.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 3.2 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 0.3 | GO:0072137 | mitral valve formation(GO:0003192) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 3.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 10.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 5.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 27.5 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 11.3 | GO:0043436 | carboxylic acid metabolic process(GO:0019752) oxoacid metabolic process(GO:0043436) |
| 0.0 | 29.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 92.0 | GO:0042627 | chylomicron(GO:0042627) |
| 14.5 | 174.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 14.1 | 182.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 3.1 | 159.7 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 2.9 | 34.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 2.3 | 38.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 1.7 | 213.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.0 | 13.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 1.0 | 4.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.9 | 2.6 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.7 | 20.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.7 | 2.1 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.7 | 8.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.7 | 4.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.6 | 40.0 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.5 | 10.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.5 | 40.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.5 | 13.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.5 | 26.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 1.9 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.4 | 27.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.4 | 3.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 15.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 2.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 35.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 5.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 5.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 5.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 18.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 46.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 6.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 110.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 42.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 6.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 119.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 202.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 4.9 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.1 | 60.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 2.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 10.4 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 7.3 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 30.7 | 92.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 16.9 | 50.8 | GO:0046911 | metal chelating activity(GO:0046911) |
| 15.9 | 63.6 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 15.6 | 62.4 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 14.1 | 225.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 13.6 | 54.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 13.1 | 117.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 11.9 | 142.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 8.0 | 40.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 6.9 | 20.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 6.1 | 18.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 5.8 | 34.7 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 4.9 | 38.8 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 3.7 | 45.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 3.7 | 11.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 3.6 | 40.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 3.5 | 17.3 | GO:0030283 | testosterone dehydrogenase [NAD(P)] activity(GO:0030283) |
| 3.0 | 53.4 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 2.9 | 11.6 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 2.5 | 297.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 2.4 | 51.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 2.2 | 91.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 2.1 | 6.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 1.8 | 34.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 1.5 | 5.9 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 1.5 | 11.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 1.4 | 35.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.4 | 27.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 1.3 | 17.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 1.2 | 7.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.1 | 6.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.0 | 4.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 1.0 | 19.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.8 | 18.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.8 | 37.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 4.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.7 | 32.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.7 | 54.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.6 | 15.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.6 | 1.7 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.5 | 4.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.4 | 6.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 1.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.4 | 9.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.4 | 3.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 1.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.3 | 1.0 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.3 | 2.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 15.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 52.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 2.7 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 37.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.2 | 3.0 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 7.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 8.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 8.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 4.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 2.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 5.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 2.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 3.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 2.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 29.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 61.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 2.1 | 35.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 1.3 | 40.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 1.3 | 54.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 1.2 | 295.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 1.1 | 38.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 1.1 | 11.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 1.0 | 158.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.9 | 20.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.9 | 34.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.3 | 13.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 14.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 7.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 6.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 8.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 185.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 8.4 | 142.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 4.4 | 61.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 4.0 | 40.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 2.7 | 23.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.1 | 13.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.0 | 18.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 1.0 | 11.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 1.0 | 17.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.8 | 33.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.8 | 40.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.8 | 23.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.8 | 141.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.8 | 17.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.7 | 34.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.6 | 13.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.6 | 40.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.6 | 11.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.6 | 9.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.6 | 8.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.5 | 18.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 4.9 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 11.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 6.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 5.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 6.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |