Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
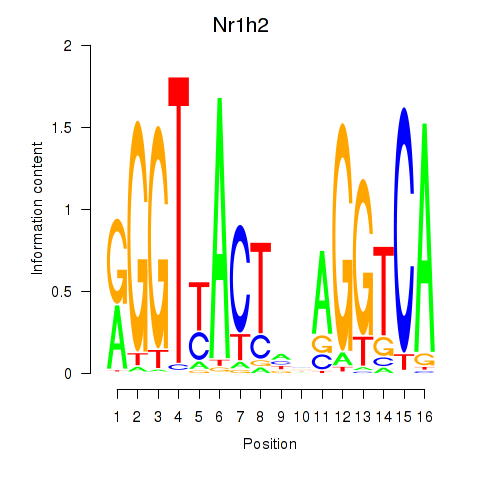
Results for Nr1h2
Z-value: 0.89
Transcription factors associated with Nr1h2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1h2
|
ENSRNOG00000019812 | nuclear receptor subfamily 1, group H, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h2 | rn6_v1_chr1_-_100559942_100559942 | -0.12 | 3.3e-02 | Click! |
Activity profile of Nr1h2 motif
Sorted Z-values of Nr1h2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_7058314 | 39.14 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr15_+_33632416 | 35.30 |
ENSRNOT00000068212
|
AC130940.1
|
|
| chr9_+_27402381 | 31.59 |
ENSRNOT00000077372
|
Gsta3
|
glutathione S-transferase alpha 3 |
| chr10_-_62273119 | 31.05 |
ENSRNOT00000004322
|
Serpinf2
|
serpin family F member 2 |
| chr2_+_60337667 | 28.06 |
ENSRNOT00000024035
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr13_-_82758004 | 26.68 |
ENSRNOT00000003932
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr1_+_224824799 | 26.39 |
ENSRNOT00000024757
|
Slc22a6
|
solute carrier family 22 member 6 |
| chr16_-_81797815 | 24.86 |
ENSRNOT00000026666
|
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr4_+_122365093 | 24.81 |
ENSRNOT00000024011
|
Klf15
|
Kruppel-like factor 15 |
| chr12_+_9446940 | 22.32 |
ENSRNOT00000074791
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5-) decarboxylase |
| chr1_-_162385575 | 19.72 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr3_+_150910398 | 18.47 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr3_+_8832740 | 17.43 |
ENSRNOT00000022552
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr4_-_117790350 | 17.20 |
ENSRNOT00000088557
|
Nat8b
|
N-acetyltransferase 8B |
| chr1_+_282557426 | 16.93 |
ENSRNOT00000088966
|
AABR07007134.1
|
|
| chr1_+_100161872 | 16.69 |
ENSRNOT00000025777
|
Klk1c3
|
kallikrein 1-related peptidase C3 |
| chr5_+_4982348 | 16.52 |
ENSRNOT00000010369
|
Lactb2
|
lactamase, beta 2 |
| chr2_-_100372252 | 16.41 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr4_-_117607428 | 16.19 |
ENSRNOT00000021243
|
LOC103690139
|
probable N-acetyltransferase CML6 |
| chr8_+_50537009 | 15.51 |
ENSRNOT00000080658
|
Apoa4
|
apolipoprotein A4 |
| chr20_+_20236151 | 13.84 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr4_-_145390447 | 13.24 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr2_-_140464607 | 13.11 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr4_+_51614676 | 12.62 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr5_-_138470096 | 12.08 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr3_+_98297554 | 11.84 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr5_-_169212170 | 11.62 |
ENSRNOT00000013385
|
Tas1r1
|
taste 1 receptor member 1 |
| chrX_+_68752597 | 11.38 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr20_+_21316826 | 11.16 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr9_+_81631409 | 10.63 |
ENSRNOT00000089580
|
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr19_-_10653800 | 10.43 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr13_-_111765944 | 10.17 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr14_-_89238094 | 9.96 |
ENSRNOT00000006818
|
Sun3
|
Sad1 and UNC84 domain containing 3 |
| chr1_+_190072420 | 9.50 |
ENSRNOT00000037026
|
Abca15
|
ATP-binding cassette, subfamily A (ABC1), member 15 |
| chr10_-_56558487 | 9.31 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr20_+_13827660 | 9.26 |
ENSRNOT00000093131
|
Ddt
|
D-dopachrome tautomerase |
| chr5_+_120340646 | 9.14 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr3_-_94808861 | 9.00 |
ENSRNOT00000038464
|
Prrg4
|
proline rich and Gla domain 4 |
| chr10_-_46582854 | 8.73 |
ENSRNOT00000004753
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr1_-_188407132 | 8.70 |
ENSRNOT00000073233
|
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr9_-_60255746 | 8.36 |
ENSRNOT00000093710
|
Dnah7
|
dynein, axonemal, heavy chain 7 |
| chr9_+_16647598 | 8.02 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr12_-_38995570 | 7.48 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr10_-_82785142 | 7.47 |
ENSRNOT00000005381
|
Sgca
|
sarcoglycan, alpha |
| chr9_+_64066579 | 7.33 |
ENSRNOT00000013511
|
RGD1306941
|
similar to CG31122-PA |
| chr9_-_16647458 | 7.24 |
ENSRNOT00000024380
|
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr1_-_70235091 | 7.03 |
ENSRNOT00000075745
|
Apeg3
|
antisense paternally expressed gene 3 |
| chr7_+_141034982 | 7.00 |
ENSRNOT00000091050
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr5_+_138470069 | 6.84 |
ENSRNOT00000076343
ENSRNOT00000064073 |
Zmynd12
|
zinc finger, MYND-type containing 12 |
| chr1_+_140477868 | 6.41 |
ENSRNOT00000025008
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr1_-_220786615 | 6.35 |
ENSRNOT00000038198
|
Tsga10ip
|
testis specific 10 interacting protein |
| chr9_-_10047507 | 6.18 |
ENSRNOT00000072118
|
Alkbh7
|
alkB homolog 7 |
| chr6_+_76675418 | 6.05 |
ENSRNOT00000076169
ENSRNOT00000010948 ENSRNOT00000082286 |
Brms1l
Brms1l
|
breast cancer metastasis-suppressor 1-like breast cancer metastasis-suppressor 1-like |
| chrX_+_26294066 | 6.01 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr5_-_78376032 | 5.83 |
ENSRNOT00000075916
|
Alad
|
aminolevulinate dehydratase |
| chr6_+_99444013 | 5.53 |
ENSRNOT00000058642
|
Ppp1r36
|
protein phosphatase 1, regulatory subunit 36 |
| chr10_+_15485905 | 5.13 |
ENSRNOT00000027609
|
Tmem8a
|
transmembrane protein 8A |
| chr5_+_117698764 | 5.07 |
ENSRNOT00000011486
|
Angptl3
|
angiopoietin-like 3 |
| chr3_-_123206828 | 4.98 |
ENSRNOT00000030192
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr1_+_88182155 | 4.95 |
ENSRNOT00000083176
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chrX_-_72515057 | 4.75 |
ENSRNOT00000076850
ENSRNOT00000041138 |
Phka1
|
phosphorylase kinase, alpha 1 |
| chr10_-_86355006 | 4.70 |
ENSRNOT00000075076
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr12_+_41543696 | 4.65 |
ENSRNOT00000079639
|
Tpcn1
|
two pore segment channel 1 |
| chr14_+_80195715 | 4.65 |
ENSRNOT00000010784
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr12_+_45905371 | 4.40 |
ENSRNOT00000039275
|
Hspb8
|
heat shock protein family B (small) member 8 |
| chr7_-_129919946 | 4.30 |
ENSRNOT00000079976
|
AABR07058658.1
|
|
| chr1_-_153096269 | 4.27 |
ENSRNOT00000022578
|
Tmem135
|
transmembrane protein 135 |
| chr1_-_80689171 | 4.03 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr15_-_32925673 | 3.90 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chrX_-_72515320 | 3.70 |
ENSRNOT00000076725
|
Phka1
|
phosphorylase kinase, alpha 1 |
| chr5_+_157423213 | 3.46 |
ENSRNOT00000023431
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr15_+_8739589 | 3.39 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr1_+_88182585 | 3.32 |
ENSRNOT00000044145
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chrX_-_13116743 | 3.30 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr3_+_123754057 | 3.29 |
ENSRNOT00000034201
ENSRNOT00000084671 |
Ap5s1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr7_-_144269486 | 3.27 |
ENSRNOT00000090051
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr4_+_156482446 | 3.22 |
ENSRNOT00000039327
|
Vom2r50
|
vomeronasal 2 receptor, 50 |
| chr8_+_119121669 | 3.21 |
ENSRNOT00000028479
|
Prss46
|
protease, serine, 46 |
| chr10_-_14061703 | 3.21 |
ENSRNOT00000017917
|
Gfer
|
growth factor, augmenter of liver regeneration |
| chr1_+_87180624 | 3.19 |
ENSRNOT00000056933
ENSRNOT00000027987 |
LOC103689986
|
protein YIF1B |
| chr16_-_70705128 | 3.14 |
ENSRNOT00000071258
|
Rnf170
|
ring finger protein 170 |
| chr3_+_114869459 | 3.09 |
ENSRNOT00000056083
|
Bloc1s6
|
biogenesis of lysosomal organelles complex 1 subunit 6 |
| chr1_-_175657485 | 3.08 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr12_+_11347643 | 2.96 |
ENSRNOT00000038175
ENSRNOT00000075986 |
Kpna7
|
karyopherin subunit alpha 7 |
| chr16_+_47368768 | 2.95 |
ENSRNOT00000017966
|
Wwc2
|
WW and C2 domain containing 2 |
| chr18_-_29290447 | 2.94 |
ENSRNOT00000025353
|
Pfdn1
|
prefoldin subunit 1 |
| chr2_-_188727670 | 2.94 |
ENSRNOT00000074920
|
Lenep
|
lens epithelial protein |
| chr3_-_74738366 | 2.91 |
ENSRNOT00000012938
|
Olr490
|
olfactory receptor 490 |
| chr9_+_53626592 | 2.76 |
ENSRNOT00000031267
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr14_+_11095163 | 2.75 |
ENSRNOT00000003076
|
Tmem150c
|
transmembrane protein 150C |
| chr9_+_37462693 | 2.73 |
ENSRNOT00000016316
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr15_-_4026637 | 2.71 |
ENSRNOT00000012372
|
Chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr6_+_99625306 | 2.62 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr17_+_507377 | 2.54 |
ENSRNOT00000023638
|
Npepo
|
aminopeptidase O |
| chr6_-_129509720 | 2.52 |
ENSRNOT00000006131
|
Atg2b
|
autophagy related 2B |
| chr12_-_49412796 | 2.48 |
ENSRNOT00000092480
|
Tmem211
|
transmembrane protein 211 |
| chr7_-_144837395 | 2.39 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr13_-_99982358 | 2.17 |
ENSRNOT00000004114
|
LOC690271
|
similar to mitochondrial ribosomal protein S11 |
| chr17_-_24182425 | 2.15 |
ENSRNOT00000024206
|
Mcur1
|
mitochondrial calcium uniporter regulator 1 |
| chr20_+_5100415 | 2.14 |
ENSRNOT00000001122
|
Ly6g5c
|
lymphocyte antigen 6 complex, locus G5C |
| chr1_-_140477796 | 2.14 |
ENSRNOT00000067172
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr5_+_141530211 | 2.14 |
ENSRNOT00000056546
|
4933427I04Rik
|
Riken cDNA 4933427I04 gene |
| chr4_+_1566448 | 2.12 |
ENSRNOT00000088227
|
Olr1243
|
olfactory receptor 1243 |
| chr1_+_172728264 | 2.10 |
ENSRNOT00000075297
|
Olr267
|
olfactory receptor 267 |
| chr4_+_157181795 | 2.07 |
ENSRNOT00000017090
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr3_-_73178585 | 2.05 |
ENSRNOT00000043965
|
Olr459
|
olfactory receptor 459 |
| chr8_+_42017493 | 2.04 |
ENSRNOT00000072062
|
LOC100911845
|
olfactory receptor 143-like |
| chr9_+_99763768 | 2.03 |
ENSRNOT00000049910
|
Olr1351
|
olfactory receptor 1351 |
| chr1_+_214562897 | 1.96 |
ENSRNOT00000085125
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr5_+_69784806 | 1.96 |
ENSRNOT00000024722
|
Olr853
|
olfactory receptor 853 |
| chr3_-_73124644 | 1.93 |
ENSRNOT00000012711
|
Olr455
|
olfactory receptor 455 |
| chr5_+_144281614 | 1.84 |
ENSRNOT00000014383
|
Trappc3
|
trafficking protein particle complex 3 |
| chr1_+_87790104 | 1.83 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr10_+_49368314 | 1.81 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr9_+_99776886 | 1.80 |
ENSRNOT00000056602
|
Olr1352
|
olfactory receptor 1352 |
| chr3_+_73138229 | 1.77 |
ENSRNOT00000092148
|
Olr456
|
olfactory receptor 456 |
| chr20_+_4188766 | 1.75 |
ENSRNOT00000081438
ENSRNOT00000060378 |
Tesb
|
testis specific basic protein |
| chr6_+_101288951 | 1.68 |
ENSRNOT00000046901
|
RGD1562540
|
RGD1562540 |
| chr4_+_59365399 | 1.68 |
ENSRNOT00000034929
|
LOC689042
|
hypothetical protein LOC689042 |
| chr2_+_188561429 | 1.66 |
ENSRNOT00000076458
ENSRNOT00000027867 |
Krtcap2
|
keratinocyte associated protein 2 |
| chr10_-_89084885 | 1.65 |
ENSRNOT00000027452
|
Plekhh3
|
pleckstrin homology, MyTH4 and FERM domain containing H3 |
| chr2_-_137404996 | 1.63 |
ENSRNOT00000090907
|
LOC365791
|
similar to T-cell activation Rho GTPase-activating protein isoform b |
| chr2_+_30685840 | 1.58 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr4_+_87608301 | 1.57 |
ENSRNOT00000058702
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr4_+_119465253 | 1.53 |
ENSRNOT00000080213
|
Vom1r102
|
vomeronasal 1 receptor 102 |
| chr4_+_1625754 | 1.51 |
ENSRNOT00000075645
|
Olr1248
|
olfactory receptor 1248 |
| chr12_+_18516946 | 1.51 |
ENSRNOT00000029485
|
Dhrsx
|
dehydrogenase/reductase X-linked |
| chr8_+_42549930 | 1.46 |
ENSRNOT00000050959
|
Olr1260
|
olfactory receptor 1260 |
| chr3_+_80555196 | 1.43 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr3_-_5330466 | 1.37 |
ENSRNOT00000081193
|
AABR07051348.1
|
|
| chr12_+_10255416 | 1.37 |
ENSRNOT00000092720
|
Gpr12
|
G protein-coupled receptor 12 |
| chr1_-_172943853 | 1.35 |
ENSRNOT00000047040
|
Olr278
|
olfactory receptor 278 |
| chr3_+_11476883 | 1.24 |
ENSRNOT00000072241
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr8_+_42338410 | 1.24 |
ENSRNOT00000073541
|
Olr1246
|
olfactory receptor 1246 |
| chr4_-_161658519 | 1.22 |
ENSRNOT00000007634
ENSRNOT00000067895 |
Tulp3
|
tubby-like protein 3 |
| chr20_+_28920616 | 1.18 |
ENSRNOT00000070897
|
P4ha1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr17_-_66511870 | 1.14 |
ENSRNOT00000044662
|
Lgals8
|
galectin 8 |
| chr8_+_61901128 | 1.12 |
ENSRNOT00000033209
|
LOC691110
|
similar to taste receptor protein 1 |
| chr10_-_104163634 | 1.10 |
ENSRNOT00000005198
|
Mif4gd
|
MIF4G domain containing |
| chr1_-_172486268 | 1.10 |
ENSRNOT00000083135
|
Olr255
|
olfactory receptor 255 |
| chr4_+_1436254 | 1.08 |
ENSRNOT00000088564
|
Olr1232
|
olfactory receptor 1232 |
| chr3_-_158936972 | 1.03 |
ENSRNOT00000074733
|
Olr1254
|
olfactory receptor 1254 |
| chr4_+_1601652 | 1.00 |
ENSRNOT00000072284
|
LOC100912152
|
olfactory receptor 8B3-like |
| chr7_-_58286770 | 0.98 |
ENSRNOT00000005258
|
Rab21
|
RAB21, member RAS oncogene family |
| chr11_+_61531416 | 0.91 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chrX_+_92596378 | 0.90 |
ENSRNOT00000045001
|
Pcdh11x
|
protocadherin 11 X-linked |
| chr20_-_32628953 | 0.89 |
ENSRNOT00000000451
|
Gprc6a
|
G protein-coupled receptor, class C, group 6, member A |
| chrX_-_43781586 | 0.82 |
ENSRNOT00000051551
|
Cldn34b
|
claudin 34B |
| chr8_+_12284528 | 0.81 |
ENSRNOT00000008566
|
Mtmr2
|
myotubularin related protein 2 |
| chr1_+_63333283 | 0.79 |
ENSRNOT00000086294
|
Vom1r30
|
vomeronasal 1 receptor 30 |
| chr11_-_28900376 | 0.73 |
ENSRNOT00000061606
|
Krtap16-5
|
keratin associated protein 16-5 |
| chr20_+_1197746 | 0.70 |
ENSRNOT00000045794
|
Olr1710
|
olfactory receptor 1710 |
| chr11_+_61531571 | 0.68 |
ENSRNOT00000093467
ENSRNOT00000002727 |
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr3_-_73401074 | 0.68 |
ENSRNOT00000012848
|
Olr476
|
olfactory receptor 476 |
| chr3_+_51687809 | 0.67 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr14_+_79401834 | 0.66 |
ENSRNOT00000082704
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr3_-_151600140 | 0.64 |
ENSRNOT00000026451
|
Fer1l4
|
fer-1-like family member 4 |
| chr3_-_103043741 | 0.61 |
ENSRNOT00000052294
|
Olr775
|
olfactory receptor 775 |
| chr1_-_172465446 | 0.61 |
ENSRNOT00000088386
|
LOC103690410
|
olfactory receptor 484-like |
| chr3_+_38742072 | 0.55 |
ENSRNOT00000006774
|
Arl6ip6
|
ADP ribosylation factor like GTPase 6 interacting protein 6 |
| chr6_-_38007162 | 0.54 |
ENSRNOT00000044583
|
AABR07063632.1
|
|
| chr4_-_97567844 | 0.50 |
ENSRNOT00000042169
|
Vom1r89
|
vomeronasal 1 receptor 89 |
| chr6_-_105160470 | 0.48 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr18_-_33414236 | 0.47 |
ENSRNOT00000020385
|
Yipf5
|
Yip1 domain family, member 5 |
| chr1_+_214664537 | 0.42 |
ENSRNOT00000075002
|
AABR07006030.1
|
|
| chr7_-_144837583 | 0.36 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr4_+_87516218 | 0.35 |
ENSRNOT00000090182
|
Vom1r66
|
vomeronasal 1 receptor 66 |
| chr7_+_120202601 | 0.34 |
ENSRNOT00000082862
|
AC096473.3
|
|
| chr5_+_136406130 | 0.33 |
ENSRNOT00000093099
|
Eri3
|
ERI1 exoribonuclease family member 3 |
| chr1_+_260798239 | 0.29 |
ENSRNOT00000036791
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr10_-_87459652 | 0.24 |
ENSRNOT00000018804
|
Krt40
|
keratin 40 |
| chr4_-_121811539 | 0.24 |
ENSRNOT00000049591
|
Vom1r94
|
vomeronasal 1 receptor 94 |
| chr9_+_65534704 | 0.23 |
ENSRNOT00000016730
|
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chrX_+_22788660 | 0.21 |
ENSRNOT00000071886
|
AABR07037510.1
|
|
| chr8_+_42347811 | 0.04 |
ENSRNOT00000073897
|
LOC100912186
|
olfactory receptor 8B3-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1h2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.5 | 31.6 | GO:0046223 | toxin catabolic process(GO:0009407) mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 9.8 | 39.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 9.4 | 28.1 | GO:0019265 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) |
| 8.9 | 26.7 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 6.2 | 31.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 5.2 | 15.5 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 4.4 | 26.4 | GO:0031427 | response to methotrexate(GO:0031427) |
| 3.5 | 13.8 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.3 | 7.0 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 2.3 | 9.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 2.1 | 10.6 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 2.1 | 6.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 1.9 | 11.6 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 1.7 | 10.4 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 1.7 | 5.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 1.3 | 5.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 1.2 | 10.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 1.1 | 28.5 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 1.0 | 13.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 1.0 | 5.8 | GO:0010266 | response to vitamin B1(GO:0010266) response to platinum ion(GO:0070541) |
| 0.9 | 24.8 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.8 | 8.4 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.7 | 12.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.7 | 26.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.6 | 3.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.5 | 9.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.5 | 3.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.5 | 8.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 24.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.4 | 0.8 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.4 | 7.5 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.4 | 9.3 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.4 | 7.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.3 | 3.1 | GO:0043476 | endosome to melanosome transport(GO:0035646) pigment accumulation(GO:0043476) cellular pigment accumulation(GO:0043482) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.3 | 1.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.3 | 1.4 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.2 | 3.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 | 6.0 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.2 | 1.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 24.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 6.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 14.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 2.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 9.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 4.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 2.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.2 | 10.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 3.4 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.2 | 3.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 4.9 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 0.7 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 2.7 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.1 | 4.7 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.1 | 1.0 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 10.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.2 | GO:1903943 | negative regulation of myoblast fusion(GO:1901740) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 2.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 6.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.7 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.1 | 6.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 3.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 11.8 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.8 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 1.2 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 3.3 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 33.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 4.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 5.0 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.0 | 2.1 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.6 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 6.9 | GO:0016567 | protein ubiquitination(GO:0016567) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 31.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.2 | 26.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 2.1 | 8.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 1.4 | 10.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.4 | 13.8 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.2 | 7.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.2 | 15.5 | GO:0042627 | chylomicron(GO:0042627) |
| 1.1 | 8.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 1.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.7 | 9.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.6 | 17.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.5 | 17.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.4 | 2.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 1.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.3 | 1.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.3 | 6.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 26.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.3 | 13.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 3.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 26.4 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 6.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 3.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 62.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 11.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 2.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 8.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 11.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 3.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 49.7 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 11.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 3.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 4.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 3.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.5 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 28.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 3.3 | 26.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 3.1 | 9.3 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 2.9 | 8.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 2.5 | 17.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 2.2 | 15.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 2.1 | 10.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 2.0 | 38.5 | GO:0030955 | potassium ion binding(GO:0030955) |
| 1.9 | 5.8 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 1.8 | 7.0 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.7 | 8.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.4 | 39.1 | GO:0019825 | oxygen binding(GO:0019825) |
| 1.4 | 8.7 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.1 | 3.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 1.0 | 9.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.9 | 4.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.9 | 6.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.8 | 31.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.7 | 6.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.6 | 12.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.5 | 22.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.5 | 2.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.4 | 7.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 11.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.3 | 24.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.3 | 3.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.3 | 10.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.3 | 14.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.3 | 31.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 44.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 9.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 16.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 11.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 5.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 5.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 6.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 3.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 18.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 10.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 12.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 2.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 4.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 3.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.6 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 2.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 17.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 4.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 2.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 5.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 16.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 9.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 32.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 7.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 9.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 5.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 39.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.5 | 24.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 2.4 | 26.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 2.2 | 28.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.1 | 31.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.9 | 12.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.9 | 26.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.6 | 7.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.6 | 9.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.6 | 16.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.6 | 12.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 8.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 9.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 5.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 31.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 10.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 8.7 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.3 | 8.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 11.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 13.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 12.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 12.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |