Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
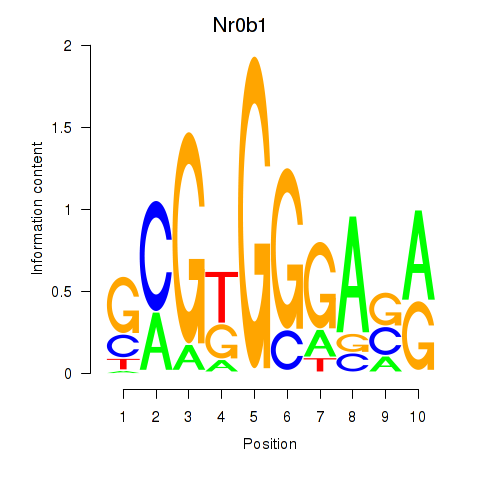
Results for Nr0b1
Z-value: 0.25
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSRNOG00000003765 | nuclear receptor subfamily 0, group B, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | rn6_v1_chrX_+_54734385_54734385 | 0.17 | 2.7e-03 | Click! |
Activity profile of Nr0b1 motif
Sorted Z-values of Nr0b1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_83471342 | 10.34 |
ENSRNOT00000019384
|
Neto1
|
neuropilin and tolloid like 1 |
| chr12_+_31530699 | 8.58 |
ENSRNOT00000033379
ENSRNOT00000067867 |
Rimbp2
|
RIMS binding protein 2 |
| chr3_-_39596718 | 4.98 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr2_-_186245342 | 4.07 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr7_+_80351774 | 3.78 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
| chr6_+_108167716 | 3.52 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr2_-_84531192 | 3.15 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr5_+_135074297 | 3.14 |
ENSRNOT00000000157
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr3_+_97721668 | 2.67 |
ENSRNOT00000065168
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr8_-_50277797 | 2.46 |
ENSRNOT00000082508
|
Pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr8_+_49441106 | 2.19 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr9_+_95221474 | 1.97 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chrX_-_134866210 | 1.92 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr2_+_113066885 | 1.68 |
ENSRNOT00000034960
|
Ghsr
|
growth hormone secretagogue receptor |
| chr9_+_66568074 | 1.41 |
ENSRNOT00000035238
|
Bmpr2
|
bone morphogenetic protein receptor type 2 |
| chr19_-_26194198 | 1.31 |
ENSRNOT00000005917
|
Wdr83
|
WD repeat domain 83 |
| chr3_+_153398130 | 0.57 |
ENSRNOT00000068135
|
Rpn2
|
ribophorin II |
| chr10_-_6870011 | 0.45 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
| chr4_-_156427755 | 0.13 |
ENSRNOT00000085675
ENSRNOT00000014030 |
LOC103690024
|
peroxisomal targeting signal 1 receptor-like |
| chr4_-_157008947 | 0.09 |
ENSRNOT00000073341
|
Pex5
|
peroxisomal biogenesis factor 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr0b1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.6 | 1.7 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.5 | 1.9 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.5 | 1.4 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) regulation of lung blood pressure(GO:0014916) negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) |
| 0.4 | 3.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 2.0 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 4.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 3.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 2.2 | GO:0086027 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.1 | 3.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 8.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 2.5 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 5.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.5 | 3.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 3.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 4.1 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.6 | 1.7 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.5 | 3.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 2.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 1.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.2 | 10.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |