Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
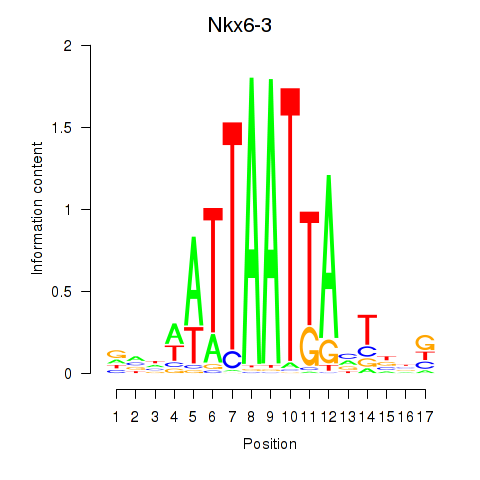
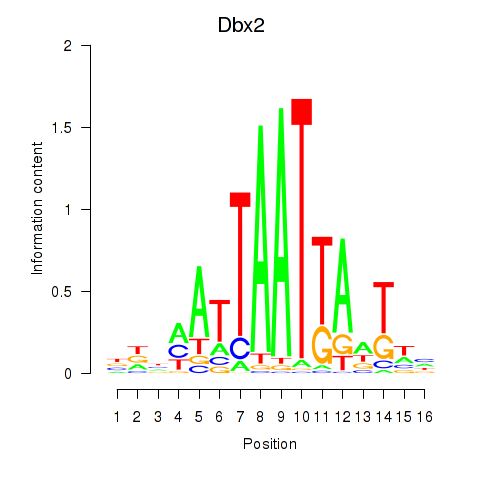
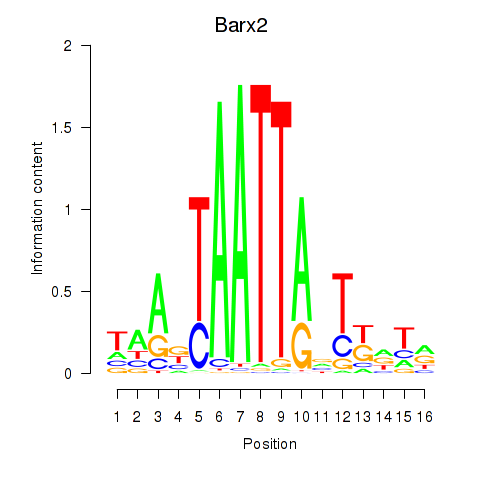
Results for Nkx6-3_Dbx2_Barx2
Z-value: 0.72
Transcription factors associated with Nkx6-3_Dbx2_Barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-3
|
ENSRNOG00000018147 | NK6 homeobox 3 |
|
Dbx2
|
ENSRNOG00000006885 | developing brain homeobox 2 |
|
Barx2
|
ENSRNOG00000008592 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barx2 | rn6_v1_chr8_-_33017854_33017854 | -0.42 | 8.9e-15 | Click! |
| Nkx6-3 | rn6_v1_chr16_-_73670482_73670482 | -0.37 | 4.6e-12 | Click! |
| Dbx2 | rn6_v1_chr7_-_136997050_136997050 | 0.07 | 2.4e-01 | Click! |
Activity profile of Nkx6-3_Dbx2_Barx2 motif
Sorted Z-values of Nkx6-3_Dbx2_Barx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_71139267 | 20.66 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr4_+_57925323 | 19.69 |
ENSRNOT00000085798
|
Cpa5
|
carboxypeptidase A5 |
| chr2_+_88217188 | 16.41 |
ENSRNOT00000014267
|
Car1
|
carbonic anhydrase I |
| chr12_-_46493203 | 14.67 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr16_+_2634603 | 14.14 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr9_+_73418607 | 14.03 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr10_+_103206014 | 13.69 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr2_-_195678848 | 11.74 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr6_-_86223052 | 11.58 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr9_+_73378057 | 11.23 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr14_+_23405717 | 10.86 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr3_-_127500709 | 10.84 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr1_+_101603222 | 10.71 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr8_-_113689681 | 10.34 |
ENSRNOT00000056435
|
LOC688828
|
similar to Nucleoside diphosphate-linked moiety X motif 16 (Nudix motif 16) |
| chr4_+_162077188 | 9.30 |
ENSRNOT00000090431
|
LOC100910725
|
C-type lectin domain family 2 member D-related protein-like |
| chr16_-_24951612 | 9.25 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr4_+_52199416 | 9.23 |
ENSRNOT00000009537
|
Spam1
|
sperm adhesion molecule 1 |
| chr6_+_64789940 | 9.06 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr10_-_87564327 | 8.52 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr9_-_85243001 | 8.10 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr1_+_185863043 | 7.71 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr1_-_266428239 | 7.69 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr6_-_140880070 | 7.43 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr4_-_168656673 | 7.35 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr1_+_105285419 | 7.31 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chrX_+_131381134 | 7.29 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr5_-_162751128 | 7.21 |
ENSRNOT00000068281
|
RGD1559644
|
similar to novel protein similar to esterases |
| chr7_-_2431197 | 7.20 |
ENSRNOT00000003498
|
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr3_+_55461420 | 6.51 |
ENSRNOT00000073549
|
G6pc2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr14_+_81858737 | 6.49 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr5_-_59165160 | 6.19 |
ENSRNOT00000029035
|
Fam221b
|
family with sequence similarity 221, member B |
| chr16_+_68586235 | 6.12 |
ENSRNOT00000039592
|
LOC103693984
|
uncharacterized LOC103693984 |
| chr12_-_35979193 | 6.04 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr1_-_227457629 | 5.99 |
ENSRNOT00000035910
ENSRNOT00000073770 |
Ms4a5
|
membrane spanning 4-domains A5 |
| chr1_+_140998240 | 5.91 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr2_-_29248174 | 5.90 |
ENSRNOT00000071167
|
LOC684444
|
similar to HIStone family member (his-41) |
| chrX_+_144994139 | 5.89 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr4_-_69196430 | 5.77 |
ENSRNOT00000017673
|
LOC312273
|
Trypsin V-A |
| chr10_-_87286387 | 5.73 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr8_-_12993651 | 5.73 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr10_+_87759769 | 5.64 |
ENSRNOT00000017378
ENSRNOT00000046526 |
Krtap9-1
|
keratin associated protein 9-1 |
| chr13_+_49005405 | 5.55 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr16_+_75572070 | 5.54 |
ENSRNOT00000043486
|
Defb52
|
defensin beta 52 |
| chr16_+_72010106 | 5.49 |
ENSRNOT00000058330
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr7_+_44009069 | 5.18 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr14_-_33444235 | 5.12 |
ENSRNOT00000044441
|
Arl9
|
ADP-ribosylation factor like GTPase 9 |
| chr1_+_201337416 | 4.88 |
ENSRNOT00000067742
|
Btbd16
|
BTB domain containing 16 |
| chr19_+_52258947 | 4.76 |
ENSRNOT00000021072
|
Adad2
|
adenosine deaminase domain containing 2 |
| chr3_-_15278645 | 4.73 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr6_-_142353308 | 4.73 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chr5_-_115387377 | 4.65 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr1_+_198089446 | 4.59 |
ENSRNOT00000026146
|
Sgf29
|
SAGA complex associated factor 29 |
| chr1_-_14117021 | 4.46 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr19_-_37528011 | 4.45 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr9_-_18371856 | 4.43 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr3_+_70327193 | 4.22 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr14_+_39964588 | 4.10 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr7_-_14435967 | 4.08 |
ENSRNOT00000074801
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr2_-_170301348 | 4.08 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr4_-_170740274 | 4.03 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr6_+_2216623 | 3.96 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr5_-_17061361 | 3.91 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr1_-_107373807 | 3.75 |
ENSRNOT00000056024
|
Svip
|
small VCP interacting protein |
| chr4_-_80395502 | 3.74 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chrX_+_105011489 | 3.74 |
ENSRNOT00000085068
|
Arl13a
|
ADP ribosylation factor like GTPase 13A |
| chr6_+_139405966 | 3.72 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr2_+_220298245 | 3.53 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr8_-_49308806 | 3.53 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr9_+_111233147 | 3.42 |
ENSRNOT00000056439
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr3_+_18315320 | 3.41 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr8_+_104106740 | 3.40 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr2_+_239415046 | 3.33 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr10_-_88670430 | 3.33 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr10_-_87232723 | 3.32 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr14_+_6221085 | 3.31 |
ENSRNOT00000068215
|
AABR07014259.1
|
|
| chr13_+_56546021 | 3.29 |
ENSRNOT00000016797
|
Aspm
|
abnormal spindle microtubule assembly |
| chr6_-_36876805 | 3.27 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr1_+_255040426 | 3.22 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chr2_-_183128932 | 3.13 |
ENSRNOT00000031179
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr4_+_117962319 | 3.06 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr12_+_49665282 | 3.02 |
ENSRNOT00000086397
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr4_-_138830117 | 2.83 |
ENSRNOT00000008403
|
Il5ra
|
interleukin 5 receptor subunit alpha |
| chr1_+_188420461 | 2.82 |
ENSRNOT00000037870
|
Ccp110
|
centriolar coiled-coil protein 110 |
| chr19_-_49448072 | 2.81 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr7_-_69982592 | 2.80 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr4_+_14001761 | 2.78 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chr5_+_135574172 | 2.72 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr1_-_755645 | 2.66 |
ENSRNOT00000073546
|
Vom2r6
|
vomeronasal 2 receptor, 6 |
| chr10_-_88000423 | 2.64 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr8_-_126390801 | 2.63 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr1_+_107344904 | 2.61 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chr2_+_195996521 | 2.55 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chrX_-_110230610 | 2.53 |
ENSRNOT00000093401
|
Serpina7
|
serpin family A member 7 |
| chr8_-_43304560 | 2.52 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr15_-_80713153 | 2.50 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr11_+_80358211 | 2.48 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr2_-_256154584 | 2.48 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr9_+_66058047 | 2.45 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr5_-_17061837 | 2.42 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr14_-_82171480 | 2.37 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr12_+_18679789 | 2.37 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr3_-_161294125 | 2.33 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chr4_+_14070553 | 2.32 |
ENSRNOT00000077505
|
Cd36
|
CD36 molecule |
| chr13_+_67545430 | 2.30 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr1_-_101819478 | 2.27 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr17_+_25082056 | 2.24 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr2_-_181900856 | 2.24 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr2_-_198706428 | 2.24 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr3_+_17889972 | 2.16 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr14_-_21758788 | 2.15 |
ENSRNOT00000038520
|
2310003L06Rik
|
RIKEN cDNA 2310003L06 gene |
| chr3_+_19366370 | 2.15 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr3_-_165537940 | 2.11 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr4_+_14212925 | 2.04 |
ENSRNOT00000076946
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr6_-_140485913 | 2.03 |
ENSRNOT00000048463
|
AABR07065768.1
|
|
| chr10_-_16752205 | 1.93 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chr3_-_103217528 | 1.85 |
ENSRNOT00000074456
|
Olr784
|
olfactory receptor 784 |
| chr14_+_76732650 | 1.84 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr15_-_28853582 | 1.84 |
ENSRNOT00000060437
|
Olr1642
|
olfactory receptor 1642 |
| chr17_-_44538780 | 1.81 |
ENSRNOT00000085743
|
AABR07027809.1
|
|
| chr2_-_157035483 | 1.80 |
ENSRNOT00000076491
|
LOC304239
|
similar to RalA binding protein 1 |
| chr6_-_140102325 | 1.72 |
ENSRNOT00000072238
|
AABR07065750.2
|
|
| chr9_+_71915421 | 1.70 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr1_+_218058405 | 1.70 |
ENSRNOT00000028365
|
Fgf19
|
fibroblast growth factor 19 |
| chr2_-_127710448 | 1.69 |
ENSRNOT00000093733
|
Mfsd8
|
major facilitator superfamily domain containing 8 |
| chr14_-_23604834 | 1.68 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
| chr16_+_24978527 | 1.65 |
ENSRNOT00000019363
|
Tma16
|
translation machinery associated 16 homolog |
| chr5_+_28485619 | 1.64 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr16_+_48513432 | 1.64 |
ENSRNOT00000044934
|
LOC685135
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr1_-_3849080 | 1.59 |
ENSRNOT00000045301
|
RGD1560633
|
similar to ribosomal protein L27a |
| chr8_+_22648323 | 1.58 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr3_+_3389612 | 1.58 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr10_+_57125961 | 1.54 |
ENSRNOT00000029230
|
Gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr10_-_34301197 | 1.54 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr4_+_93791054 | 1.54 |
ENSRNOT00000042300
|
AABR07060788.1
|
|
| chr4_-_66955732 | 1.50 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr14_+_3204390 | 1.48 |
ENSRNOT00000088078
|
Glmn
|
glomulin, FKBP associated protein |
| chr3_-_145810834 | 1.45 |
ENSRNOT00000075429
|
LOC102555217
|
zinc finger protein 120-like |
| chr3_-_103460529 | 1.44 |
ENSRNOT00000047274
|
LOC100909573
|
olfactory receptor 4F6-like |
| chr1_-_149770781 | 1.43 |
ENSRNOT00000082185
|
Olr16
|
olfactory receptor 16 |
| chr7_+_2458833 | 1.42 |
ENSRNOT00000085092
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr3_-_45169118 | 1.38 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr1_+_101599018 | 1.38 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chr4_-_106983279 | 1.36 |
ENSRNOT00000007700
|
AABR07061152.1
|
|
| chr7_-_9174330 | 1.33 |
ENSRNOT00000051063
|
Olr1060
|
olfactory receptor 1060 |
| chr2_-_33025271 | 1.32 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_-_1952111 | 1.31 |
ENSRNOT00000061140
|
Jade3
|
jade family PHD finger 3 |
| chr12_-_23727535 | 1.29 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr3_-_104018861 | 1.28 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr2_-_185852759 | 1.26 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr10_+_111770168 | 1.26 |
ENSRNOT00000074598
|
Vom2r65
|
vomeronasal 2 receptor, 65 |
| chr11_-_82366505 | 1.26 |
ENSRNOT00000041326
|
RGD1559972
|
similar to ribosomal protein L27a |
| chr3_+_125503638 | 1.25 |
ENSRNOT00000028900
|
Crls1
|
cardiolipin synthase 1 |
| chr2_-_149444548 | 1.22 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr1_-_167971151 | 1.22 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr3_+_48106099 | 1.21 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr1_+_207887209 | 1.21 |
ENSRNOT00000021359
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E |
| chr2_-_219262901 | 1.21 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr16_-_18643309 | 1.19 |
ENSRNOT00000031680
|
Dydc2
|
DPY30 domain containing 2 |
| chr10_-_87459652 | 1.19 |
ENSRNOT00000018804
|
Krt40
|
keratin 40 |
| chr7_+_130357643 | 1.18 |
ENSRNOT00000040769
|
Klhdc7b
|
kelch domain containing 7B |
| chr13_-_88307988 | 1.18 |
ENSRNOT00000003812
|
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr5_-_56567320 | 1.16 |
ENSRNOT00000066081
|
Topors
|
TOP1 binding arginine/serine rich protein |
| chr20_+_34258791 | 1.15 |
ENSRNOT00000000468
|
Slc35f1
|
solute carrier family 35, member F1 |
| chr2_+_127525285 | 1.15 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr1_-_813517 | 1.13 |
ENSRNOT00000041332
|
Vom2r5
|
vomeronasal 2 receptor, 5 |
| chr7_+_38897278 | 1.13 |
ENSRNOT00000006450
|
Epyc
|
epiphycan |
| chr8_+_107859366 | 1.12 |
ENSRNOT00000039690
|
A4gnt
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr3_+_129885826 | 1.10 |
ENSRNOT00000009743
|
Slx4ip
|
SLX4 interacting protein |
| chr7_-_143967484 | 1.09 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr9_+_71247781 | 1.07 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr15_-_93765498 | 1.05 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr5_-_106865746 | 1.04 |
ENSRNOT00000008224
|
Ifnb1
|
interferon beta 1 |
| chrX_+_136488691 | 1.03 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr15_-_46166335 | 1.02 |
ENSRNOT00000059215
|
Defb42
|
defensin beta 42 |
| chr7_-_143167772 | 1.02 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr4_-_2201749 | 1.01 |
ENSRNOT00000089327
|
Lmbr1
|
limb development membrane protein 1 |
| chr4_+_155009479 | 1.01 |
ENSRNOT00000070906
|
AC127013.1
|
|
| chr18_-_16543992 | 1.00 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr8_-_44565861 | 0.99 |
ENSRNOT00000049921
|
AABR07069997.1
|
|
| chr11_-_13999862 | 0.96 |
ENSRNOT00000045835
|
Lipi
|
lipase I |
| chr4_-_167202106 | 0.96 |
ENSRNOT00000038581
|
Tas2r140
|
taste receptor, type 2, member 140 |
| chr10_+_82032656 | 0.93 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr1_-_67094567 | 0.93 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr6_-_142418779 | 0.92 |
ENSRNOT00000072280
ENSRNOT00000065808 |
AABR07065814.1
|
|
| chr3_+_146980923 | 0.90 |
ENSRNOT00000011654
|
Nsfl1c
|
NSFL1 cofactor |
| chr2_+_88344527 | 0.89 |
ENSRNOT00000059467
|
RGD1565641
|
RGD1565641 |
| chr1_-_149859479 | 0.85 |
ENSRNOT00000052199
|
LOC100910009
|
olfactory receptor 14A2-like |
| chr10_+_67810810 | 0.85 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr10_+_90230711 | 0.83 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr4_+_156324922 | 0.83 |
ENSRNOT00000039463
|
Vom2r48
|
vomeronasal 2 receptor, 48 |
| chr3_-_5617225 | 0.83 |
ENSRNOT00000008327
|
Tmem8c
|
transmembrane protein 8C |
| chr16_-_75241303 | 0.78 |
ENSRNOT00000058056
|
Defb2
|
defensin beta 2 |
| chr5_+_50381244 | 0.78 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr4_+_82637411 | 0.78 |
ENSRNOT00000048225
|
LOC685406
|
LRRGT00062 |
| chr4_+_163349125 | 0.78 |
ENSRNOT00000084823
|
Klre1
|
killer cell lectin-like receptor, family E, member 1 |
| chr5_+_25253010 | 0.78 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr3_-_102484517 | 0.77 |
ENSRNOT00000006522
|
Olr750
|
olfactory receptor 750 |
| chr13_+_105684420 | 0.77 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr5_+_162808646 | 0.76 |
ENSRNOT00000021155
|
Dhrs3
|
dehydrogenase/reductase 3 |
| chr5_+_25725683 | 0.76 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-3_Dbx2_Barx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 2.7 | 10.8 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 2.6 | 7.7 | GO:0010034 | response to acetate(GO:0010034) |
| 2.1 | 6.3 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 2.0 | 14.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 1.9 | 5.7 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 1.5 | 7.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.5 | 19.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 1.5 | 7.3 | GO:0036233 | glycine import(GO:0036233) |
| 1.3 | 3.8 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.0 | 4.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 1.0 | 9.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.0 | 3.0 | GO:0043474 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 1.0 | 10.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.0 | 2.9 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.9 | 2.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.9 | 8.8 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.9 | 5.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.8 | 2.4 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.6 | 1.9 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.6 | 25.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.6 | 2.4 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.6 | 3.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.6 | 10.6 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.6 | 4.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.5 | 3.3 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.5 | 14.7 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.5 | 1.6 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.5 | 4.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 1.4 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.5 | 6.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.5 | 8.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.4 | 16.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.4 | 3.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.4 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 1.1 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.3 | 3.7 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.3 | 1.7 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.3 | 1.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 4.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.3 | 2.3 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 0.6 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.3 | 0.8 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.3 | 0.8 | GO:0002835 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.3 | 1.5 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.3 | 1.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 6.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 1.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 2.5 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.2 | 3.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 2.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 4.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 3.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 | 2.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 3.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 7.2 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.2 | 1.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.7 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.2 | 1.0 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.2 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 17.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.4 | GO:0090024 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.1 | 1.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 3.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 2.8 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.1 | 2.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 1.5 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 0.5 | GO:0031053 | primary miRNA processing(GO:0031053) G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 1.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 3.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.8 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 1.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 7.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 2.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 1.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 4.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.1 | 1.7 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.3 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 3.3 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 | 2.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 5.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 5.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 1.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 1.9 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 2.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 2.6 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.4 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 2.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.2 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 26.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.1 | 6.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.8 | 5.9 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.8 | 3.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 11.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.6 | 10.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 8.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 3.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.4 | 3.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 10.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 4.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.4 | 1.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 4.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 9.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 17.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 4.6 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.3 | 1.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 5.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 16.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 3.0 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 1.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 2.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 10.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 9.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 5.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 9.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 9.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 3.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 3.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 1.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 4.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 16.7 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0015489 | ornithine decarboxylase inhibitor activity(GO:0008073) polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 2.7 | 10.8 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 2.4 | 7.3 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 2.3 | 16.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 2.2 | 19.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.4 | 4.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 1.3 | 6.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.1 | 25.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.1 | 6.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.0 | 7.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.0 | 5.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 1.0 | 4.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.9 | 9.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.9 | 3.4 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.7 | 19.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 10.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 7.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.5 | 3.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.5 | 1.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.5 | 1.4 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.5 | 5.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 4.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.4 | 6.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.4 | 4.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.4 | 1.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.4 | 1.5 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.4 | 1.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 1.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.3 | 1.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 3.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 5.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 2.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 1.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 7.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 8.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 2.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 4.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 1.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 4.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 0.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 1.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 1.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.6 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 1.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 2.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 14.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 21.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 13.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 2.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 2.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 16.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 3.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.2 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 3.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 8.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 5.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 3.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 3.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 8.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 3.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 20.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 3.4 | GO:0003712 | transcription cofactor activity(GO:0003712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 26.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 13.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 4.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 3.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 4.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 6.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 11.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.7 | 4.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 3.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 5.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 8.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 12.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 5.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.2 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 6.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 10.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 11.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |