Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Nkx6-2
Z-value: 0.60
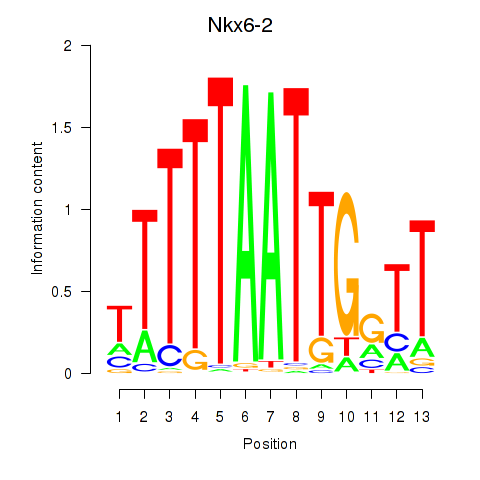
Transcription factors associated with Nkx6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-2
|
ENSRNOG00000017748 | NK6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-2 | rn6_v1_chr1_-_211923929_211923929 | 0.11 | 5.1e-02 | Click! |
Activity profile of Nkx6-2 motif
Sorted Z-values of Nkx6-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_115387377 | 17.01 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr1_+_83933942 | 11.53 |
ENSRNOT00000068690
|
Cyp2f4
|
cytochrome P450, family 2, subfamily f, polypeptide 4 |
| chr20_+_20577561 | 11.14 |
ENSRNOT00000081113
|
Cdk1
|
cyclin-dependent kinase 1 |
| chr10_+_45289741 | 10.56 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr10_-_86930947 | 10.32 |
ENSRNOT00000081440
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr2_+_206064394 | 10.22 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr17_+_43616247 | 8.62 |
ENSRNOT00000072019
|
Hist1h3a
|
histone cluster 1, H3a |
| chr2_+_92559929 | 8.29 |
ENSRNOT00000033404
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr2_-_173800761 | 8.21 |
ENSRNOT00000039524
|
Wdr49
|
WD repeat domain 49 |
| chr2_+_206064179 | 8.20 |
ENSRNOT00000025953
|
Syt6
|
synaptotagmin 6 |
| chr8_+_23398030 | 7.08 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr19_+_25391700 | 6.79 |
ENSRNOT00000011163
|
LOC100359539
|
ribonucleotide reductase M2 polypeptide |
| chr15_+_87722221 | 6.60 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr1_-_4550768 | 6.50 |
ENSRNOT00000061954
|
Adgb
|
androglobin |
| chr20_+_32450733 | 6.22 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr14_-_43143973 | 5.93 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr3_-_60611924 | 5.73 |
ENSRNOT00000068745
|
Chn1
|
chimerin 1 |
| chr2_-_96509424 | 5.61 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr18_-_26211445 | 5.49 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr18_-_58423196 | 5.49 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr7_-_120839577 | 5.44 |
ENSRNOT00000018526
|
Dmc1
|
DNA meiotic recombinase 1 |
| chr2_-_124047044 | 5.27 |
ENSRNOT00000065384
|
Cetn4
|
centrin 4 |
| chr5_+_119097715 | 5.25 |
ENSRNOT00000045987
|
Ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr5_+_133819726 | 5.21 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chr7_+_34402738 | 5.21 |
ENSRNOT00000030985
|
Ccdc38
|
coiled-coil domain containing 38 |
| chr9_+_73378057 | 5.14 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_+_70327193 | 4.67 |
ENSRNOT00000089165
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr6_-_76552559 | 4.45 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr3_+_113918629 | 4.25 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr14_-_21299068 | 4.05 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr16_+_3817402 | 3.88 |
ENSRNOT00000015011
|
Tmem254
|
transmembrane protein 254 |
| chr14_-_72025137 | 3.74 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr7_-_122644054 | 3.67 |
ENSRNOT00000025941
|
Dnajb7
|
DnaJ heat shock protein family (Hsp40) member B7 |
| chr4_-_8502280 | 3.65 |
ENSRNOT00000036164
|
AABR07059201.1
|
|
| chr3_-_121835360 | 3.61 |
ENSRNOT00000006126
ENSRNOT00000080818 |
Il1a
|
interleukin 1 alpha |
| chr6_-_95934296 | 3.55 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr3_-_56030744 | 3.55 |
ENSRNOT00000089637
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr3_+_94549180 | 3.49 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr8_+_85259982 | 3.35 |
ENSRNOT00000010409
|
Elovl5
|
ELOVL fatty acid elongase 5 |
| chrX_+_6273733 | 3.31 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr3_-_37803112 | 3.20 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr9_-_81879211 | 3.09 |
ENSRNOT00000085140
|
Rnf25
|
ring finger protein 25 |
| chr14_-_72122158 | 2.94 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr20_+_42966140 | 2.93 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr1_-_67284864 | 2.93 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chr2_+_210880777 | 2.88 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr16_+_46731403 | 2.88 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr2_-_198706428 | 2.82 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr18_+_30904498 | 2.76 |
ENSRNOT00000026969
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr4_-_727691 | 2.73 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr12_+_39423596 | 2.67 |
ENSRNOT00000066952
|
Ift81
|
intraflagellar transport 81 |
| chr5_+_173447784 | 2.63 |
ENSRNOT00000027251
|
Tnfrsf4
|
TNF receptor superfamily member 4 |
| chr4_-_66955732 | 2.61 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr1_+_193424812 | 2.56 |
ENSRNOT00000019939
|
Aqp8
|
aquaporin 8 |
| chr8_+_79638696 | 2.54 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chrX_+_31984612 | 2.53 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr14_+_88549947 | 2.52 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr1_+_99616447 | 2.45 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr10_-_85717511 | 2.44 |
ENSRNOT00000005445
|
LOC691189
|
hypothetical protein LOC691189 |
| chr9_-_78693028 | 2.41 |
ENSRNOT00000036359
|
Abca12
|
ATP binding cassette subfamily A member 12 |
| chr2_+_243577082 | 2.38 |
ENSRNOT00000016556
|
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr4_-_135069970 | 2.30 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr20_+_27173213 | 2.25 |
ENSRNOT00000073617
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr7_+_64769089 | 2.24 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr4_+_162281574 | 2.18 |
ENSRNOT00000087850
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr3_+_8430829 | 2.16 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr4_-_163762434 | 2.14 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr1_-_14117021 | 2.12 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr8_+_115546893 | 2.10 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chrX_-_153457040 | 2.10 |
ENSRNOT00000084905
|
LOC689600
|
hypothetical protein LOC689600 |
| chrX_+_131381134 | 2.07 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr3_+_153491151 | 2.04 |
ENSRNOT00000047047
|
Manbal
|
mannosidase beta like |
| chrX_+_110016995 | 2.02 |
ENSRNOT00000093542
|
Nrk
|
Nik related kinase |
| chr4_-_51931112 | 2.02 |
ENSRNOT00000080575
|
Pot1
|
protection of telomeres 1 |
| chr10_-_43599232 | 1.94 |
ENSRNOT00000044299
|
Gemin5
|
gem (nuclear organelle) associated protein 5 |
| chr2_-_138833933 | 1.94 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr13_-_104284068 | 1.86 |
ENSRNOT00000005407
|
LOC100912365
|
uncharacterized LOC100912365 |
| chr2_-_128675408 | 1.75 |
ENSRNOT00000019256
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr17_+_9596957 | 1.73 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr15_+_56666012 | 1.73 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chrX_-_138112408 | 1.70 |
ENSRNOT00000077028
|
Frmd7
|
FERM domain containing 7 |
| chr20_+_27212724 | 1.66 |
ENSRNOT00000032600
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr15_-_38096875 | 1.65 |
ENSRNOT00000032723
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr18_+_30496318 | 1.60 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr4_-_150522351 | 1.60 |
ENSRNOT00000079053
|
Zfp9
|
zinc finger protein 9 |
| chr2_+_127459012 | 1.59 |
ENSRNOT00000093685
ENSRNOT00000072675 |
Intu
|
inturned planar cell polarity protein |
| chr6_+_100337226 | 1.52 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr10_+_65552897 | 1.46 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr7_-_144837583 | 1.41 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr7_-_11223649 | 1.39 |
ENSRNOT00000061191
|
Mfsd12
|
major facilitator superfamily domain containing 12 |
| chr9_+_20951260 | 1.36 |
ENSRNOT00000016906
|
Adgrf4
|
adhesion G protein-coupled receptor F4 |
| chr4_+_168775111 | 1.36 |
ENSRNOT00000011096
|
Ddx47
|
DEAD-box helicase 47 |
| chr16_+_90325304 | 1.35 |
ENSRNOT00000057310
|
Slc10a2
|
solute carrier family 10 member 2 |
| chr13_+_104284660 | 1.30 |
ENSRNOT00000005400
|
Dusp10
|
dual specificity phosphatase 10 |
| chr12_-_51877624 | 1.29 |
ENSRNOT00000056800
|
Chek2
|
checkpoint kinase 2 |
| chr18_-_77535564 | 1.26 |
ENSRNOT00000041380
ENSRNOT00000090265 |
Atp9b
|
ATPase phospholipid transporting 9B (putative) |
| chr5_-_128730446 | 1.22 |
ENSRNOT00000089441
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr17_-_53915076 | 1.21 |
ENSRNOT00000010836
|
Arid4b
|
AT-rich interaction domain 4B |
| chr1_+_99706780 | 1.15 |
ENSRNOT00000024758
|
Klk12
|
kallikrein related-peptidase 12 |
| chr13_-_47916185 | 1.11 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr17_+_34704616 | 1.10 |
ENSRNOT00000090706
ENSRNOT00000083674 ENSRNOT00000077110 |
Exoc2
|
exocyst complex component 2 |
| chr4_+_10748745 | 1.07 |
ENSRNOT00000046142
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr6_+_129621070 | 1.07 |
ENSRNOT00000091338
|
Papola
|
poly (A) polymerase alpha |
| chrX_-_123731294 | 0.99 |
ENSRNOT00000092574
ENSRNOT00000032618 |
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr7_+_23854846 | 0.95 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr15_-_82581916 | 0.91 |
ENSRNOT00000057891
|
RGD1560069
|
similar to ribosomal protein L27 |
| chr4_+_162292305 | 0.90 |
ENSRNOT00000010098
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr2_-_137153551 | 0.89 |
ENSRNOT00000083443
|
AABR07010476.1
|
|
| chr15_+_30525754 | 0.88 |
ENSRNOT00000086842
|
AABR07017745.2
|
|
| chr3_+_55461420 | 0.86 |
ENSRNOT00000073549
|
G6pc2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr10_-_104482838 | 0.86 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr17_+_35435079 | 0.83 |
ENSRNOT00000074800
|
Exoc2
|
exocyst complex component 2 |
| chr3_+_152402520 | 0.81 |
ENSRNOT00000027086
|
Cnbd2
|
cyclic nucleotide binding domain containing 2 |
| chr4_-_160662974 | 0.80 |
ENSRNOT00000089199
|
Tspan9
|
tetraspanin 9 |
| chr9_+_73334618 | 0.75 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr9_+_1313209 | 0.72 |
ENSRNOT00000083387
|
Tbc1d5
|
TBC1 domain family, member 5 |
| chrX_+_124631881 | 0.71 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr19_+_27603307 | 0.68 |
ENSRNOT00000074702
|
AABR07043347.1
|
|
| chr13_-_52413681 | 0.66 |
ENSRNOT00000037100
ENSRNOT00000011801 |
Nav1
|
neuron navigator 1 |
| chr13_+_88644520 | 0.64 |
ENSRNOT00000003979
|
Spata46
|
spermatogenesis associated 46 |
| chr19_-_43192208 | 0.64 |
ENSRNOT00000025179
|
Exosc6
|
exosome component 6 |
| chr10_+_38661901 | 0.64 |
ENSRNOT00000009098
|
Zcchc10
|
zinc finger CCHC-type containing 10 |
| chr9_-_10369937 | 0.60 |
ENSRNOT00000071228
ENSRNOT00000072911 ENSRNOT00000072009 |
Ranbp3
|
RAN binding protein 3 |
| chr1_-_229601032 | 0.59 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr15_-_88036354 | 0.58 |
ENSRNOT00000014747
|
Ednrb
|
endothelin receptor type B |
| chr1_-_150101177 | 0.53 |
ENSRNOT00000042931
|
Olr24
|
olfactory receptor 24 |
| chr9_+_20251521 | 0.52 |
ENSRNOT00000005535
|
LOC100911625
|
gamma-enolase-like |
| chr15_-_19575914 | 0.52 |
ENSRNOT00000043897
|
AABR07017253.1
|
|
| chr19_-_21970103 | 0.51 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr15_-_32925673 | 0.49 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chrX_+_22342308 | 0.48 |
ENSRNOT00000079909
ENSRNOT00000087945 ENSRNOT00000077528 |
Kdm5c
|
lysine demethylase 5C |
| chr10_+_104483042 | 0.46 |
ENSRNOT00000007362
|
Sap30bp
|
SAP30 binding protein |
| chr3_-_104018861 | 0.45 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr17_+_25082056 | 0.45 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr4_-_157294047 | 0.43 |
ENSRNOT00000005601
|
Eno2
|
enolase 2 |
| chr19_-_11669578 | 0.42 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr14_+_108007724 | 0.38 |
ENSRNOT00000014062
|
Xpo1
|
exportin 1 |
| chr11_+_82415995 | 0.38 |
ENSRNOT00000051772
|
AABR07034641.1
|
|
| chr9_+_53546018 | 0.36 |
ENSRNOT00000016842
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr8_-_76239995 | 0.36 |
ENSRNOT00000071321
|
LOC100912027
|
60S ribosomal protein L27-like |
| chr10_+_29033989 | 0.36 |
ENSRNOT00000088322
|
Slu7
|
SLU7 homolog, splicing factor |
| chr12_+_51878153 | 0.36 |
ENSRNOT00000056798
|
Hscb
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr5_-_102743417 | 0.31 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr1_-_150173316 | 0.31 |
ENSRNOT00000044676
|
Olr27
|
olfactory receptor 27 |
| chr13_+_67545430 | 0.30 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr20_+_41266566 | 0.22 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chrX_-_138237157 | 0.07 |
ENSRNOT00000003414
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr7_-_40315377 | 0.06 |
ENSRNOT00000091319
ENSRNOT00000036141 |
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr16_-_930527 | 0.06 |
ENSRNOT00000014787
|
Spin1
|
spindlin 1 |
| chr2_-_225025972 | 0.03 |
ENSRNOT00000091767
|
AABR07013154.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.5 | GO:0042196 | chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 3.7 | 18.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 2.1 | 6.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.7 | 10.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.6 | 11.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 1.3 | 4.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 1.2 | 3.6 | GO:1904444 | regulation of establishment of Sertoli cell barrier(GO:1904444) |
| 0.8 | 5.4 | GO:0042148 | strand invasion(GO:0042148) |
| 0.7 | 2.0 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 0.7 | 3.4 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 5.9 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.6 | 5.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.6 | 1.7 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.6 | 3.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 2.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.5 | 2.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.5 | 2.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.4 | 6.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 2.5 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.4 | 2.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.4 | 2.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 1.5 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.4 | 2.6 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.3 | 2.0 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.3 | 1.3 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.3 | 1.3 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.3 | 2.6 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 2.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.2 | 1.9 | GO:0001927 | exocyst assembly(GO:0001927) entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 2.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 2.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 1.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 0.9 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 5.8 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.2 | 1.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 0.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 1.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.6 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.2 | 4.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 18.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 2.5 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 5.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 2.1 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.6 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 11.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.6 | GO:0071051 | U4 snRNA 3'-end processing(GO:0034475) DNA deamination(GO:0045006) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.9 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.5 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.4 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 5.5 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 6.6 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 1.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 5.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 3.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 1.1 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 3.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 8.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 2.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 6.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 2.6 | 10.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.9 | 18.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.9 | 2.6 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.8 | 2.5 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.7 | 2.9 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.6 | 5.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 2.4 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.2 | 2.0 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 0.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 19.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 3.9 | GO:0019013 | viral nucleocapsid(GO:0019013) |
| 0.2 | 1.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 2.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 4.0 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 2.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 6.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 5.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 6.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 5.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.8 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 2.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 8.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.9 | 5.7 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 1.5 | 5.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.1 | 6.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 5.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.7 | 2.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.6 | 1.7 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.6 | 11.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.5 | 2.0 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.5 | 1.9 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.4 | 11.5 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.4 | 2.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.4 | 21.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 2.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 3.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 2.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 6.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 5.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 2.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.2 | 1.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 3.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 2.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 5.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.9 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 2.1 | GO:0035174 | AMP-activated protein kinase activity(GO:0004679) histone serine kinase activity(GO:0035174) |
| 0.1 | 1.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 2.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.4 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 3.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 3.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 3.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 9.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 11.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 1.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.6 | GO:0004532 | exoribonuclease activity(GO:0004532) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 7.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 13.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 3.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 8.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 2.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 2.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 4.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 2.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 5.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 3.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 2.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 4.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |