Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
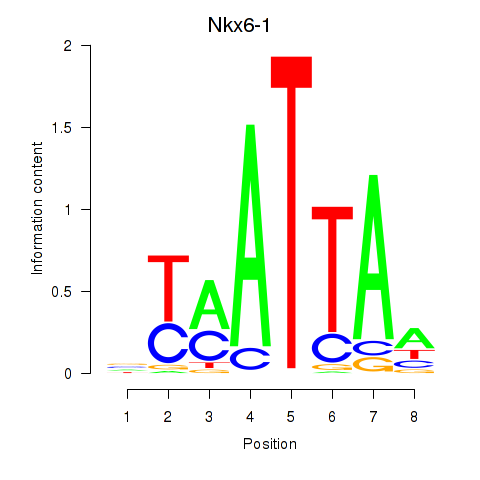
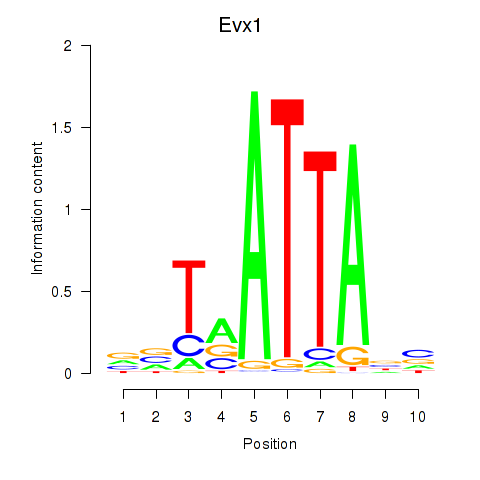
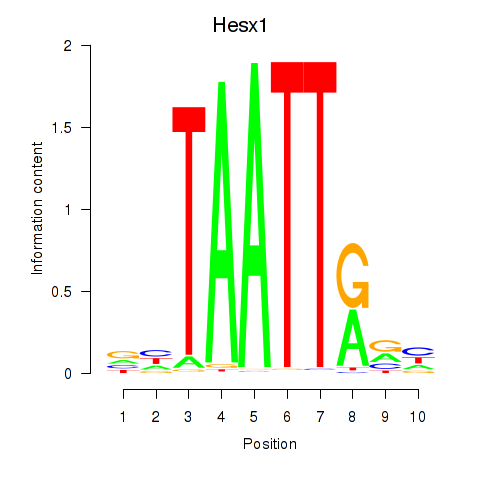
Results for Nkx6-1_Evx1_Hesx1
Z-value: 1.04
Transcription factors associated with Nkx6-1_Evx1_Hesx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-1
|
ENSRNOG00000002149 | NK6 homeobox 1 |
|
Hesx1
|
ENSRNOG00000014133 | HESX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-1 | rn6_v1_chr14_+_9555264_9555264 | 0.27 | 8.2e-07 | Click! |
| Hesx1 | rn6_v1_chr16_+_2634603_2634603 | -0.09 | 1.0e-01 | Click! |
Activity profile of Nkx6-1_Evx1_Hesx1 motif
Sorted Z-values of Nkx6-1_Evx1_Hesx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_84506328 | 55.75 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr9_-_30844199 | 50.39 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr16_+_29674793 | 40.79 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr1_+_198383201 | 36.36 |
ENSRNOT00000037405
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr3_-_52447622 | 34.74 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr7_-_69982592 | 34.29 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chrX_+_118197217 | 31.44 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr4_-_55011415 | 28.68 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr15_+_11298478 | 28.45 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr8_+_33239139 | 28.24 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr11_+_58624198 | 28.21 |
ENSRNOT00000002091
|
Gap43
|
growth associated protein 43 |
| chr17_+_63635086 | 26.56 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr7_-_76488216 | 25.80 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr2_-_45518502 | 25.36 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr6_+_58468155 | 25.29 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr17_+_11683862 | 24.75 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr2_-_158156444 | 24.59 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr3_+_48106099 | 23.25 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr1_+_13595295 | 23.01 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr3_-_63568464 | 22.87 |
ENSRNOT00000068494
|
AABR07052585.2
|
|
| chrM_+_11736 | 22.72 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr3_-_66417741 | 22.27 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr1_-_215033460 | 21.80 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr2_+_23289374 | 21.77 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr18_+_30592794 | 21.04 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr11_-_32550539 | 20.40 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chrM_+_9451 | 20.18 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr10_+_54352270 | 20.00 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chrM_+_10160 | 19.82 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_-_63836017 | 19.68 |
ENSRNOT00000030978
|
AABR07052585.1
|
|
| chr17_+_72160735 | 19.33 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr20_-_25826658 | 18.94 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr8_-_78233430 | 18.65 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr2_-_231409988 | 18.60 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr7_-_52404774 | 18.12 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr17_+_47721977 | 18.03 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr3_-_51612397 | 17.80 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr18_-_31343797 | 17.76 |
ENSRNOT00000081850
|
Pcdh1
|
protocadherin 1 |
| chr12_-_2174131 | 17.59 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr7_+_136182224 | 17.45 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr5_+_6373583 | 17.36 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr2_-_231409496 | 17.32 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chrM_+_9870 | 17.19 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr3_+_48096954 | 17.05 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr14_-_19072677 | 16.84 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr1_-_67065797 | 16.73 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr6_-_7058314 | 16.66 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chrM_+_7919 | 16.48 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr5_-_93244202 | 15.85 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr4_+_88694583 | 15.59 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr9_+_73378057 | 15.25 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_+_54253949 | 15.14 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr2_+_248398917 | 15.11 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr17_-_9762813 | 15.00 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr5_-_133959447 | 14.96 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chrM_+_7006 | 14.92 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr1_+_59156251 | 14.51 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr7_+_78558701 | 14.45 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr15_+_56666012 | 14.33 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr20_+_25990656 | 14.07 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_-_15278645 | 13.88 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr6_+_43234526 | 13.85 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr18_+_30435119 | 13.80 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr5_+_33784715 | 13.61 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr14_-_115052450 | 13.59 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr9_+_81566074 | 13.28 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr13_-_76049363 | 13.04 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr12_-_19167015 | 12.97 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chrM_+_7758 | 12.77 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_+_53044379 | 12.73 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr2_-_88763733 | 12.50 |
ENSRNOT00000059424
|
LOC688389
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr5_-_17061837 | 12.49 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr5_-_164714145 | 12.45 |
ENSRNOT00000055680
|
LOC100911485
|
mitofusin-2-like |
| chr2_-_158156150 | 11.99 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr6_+_29977797 | 11.97 |
ENSRNOT00000071784
|
Fkbp1b
|
FK506 binding protein 1B |
| chr3_-_25212049 | 11.94 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr5_-_136965191 | 11.90 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr5_-_17061361 | 11.84 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr2_+_145174876 | 11.80 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr6_-_23316962 | 11.79 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr3_+_95715193 | 11.78 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr6_+_8284878 | 11.62 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr18_-_26656879 | 11.57 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_+_84064427 | 11.55 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr13_+_87986240 | 11.54 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr3_-_45169118 | 11.51 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr11_+_80742467 | 11.42 |
ENSRNOT00000002507
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr8_-_82533689 | 11.34 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr7_+_6644643 | 11.22 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr2_+_42829413 | 11.15 |
ENSRNOT00000065528
|
Actbl2
|
actin, beta-like 2 |
| chr8_-_87315955 | 11.13 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chrX_-_142248369 | 11.05 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr13_-_93307199 | 10.95 |
ENSRNOT00000065041
|
Rgs7
|
regulator of G-protein signaling 7 |
| chr20_+_40769586 | 10.79 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr5_-_22765429 | 10.78 |
ENSRNOT00000079432
|
Asph
|
aspartate-beta-hydroxylase |
| chr16_+_2706428 | 10.77 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr18_+_17043903 | 10.50 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr18_+_59748444 | 10.48 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr16_-_10802512 | 10.42 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr10_+_90230711 | 10.42 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr10_-_87248572 | 10.38 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr12_+_47179664 | 10.38 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr18_-_53181503 | 10.35 |
ENSRNOT00000066548
|
Fbn2
|
fibrillin 2 |
| chr6_+_101532518 | 10.29 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr20_+_34633157 | 10.29 |
ENSRNOT00000000469
|
Pln
|
phospholamban |
| chr16_+_54319377 | 10.23 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr3_+_56125924 | 10.20 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr10_-_83898527 | 10.11 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr3_-_51643140 | 10.10 |
ENSRNOT00000006646
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr4_-_176909075 | 10.09 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr1_+_217345545 | 10.08 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr2_-_35104963 | 10.08 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr7_-_8060373 | 9.94 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr18_+_14471213 | 9.70 |
ENSRNOT00000045224
|
Dtna
|
dystrobrevin, alpha |
| chr16_-_49820235 | 9.65 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr17_-_84247038 | 9.59 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chrM_+_8599 | 9.53 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr10_+_56445647 | 9.48 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr16_+_10267482 | 9.27 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr10_-_91661558 | 9.23 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr8_+_57983556 | 9.21 |
ENSRNOT00000009562
|
RGD1311251
|
similar to RIKEN cDNA 4930550C14 |
| chr3_-_102672294 | 9.14 |
ENSRNOT00000030909
|
Olr760
|
olfactory receptor 760 |
| chr3_-_101547478 | 9.07 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chrX_-_32153794 | 9.06 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chrM_+_2740 | 9.01 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr12_+_18679789 | 9.00 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr10_-_51778939 | 8.96 |
ENSRNOT00000078675
ENSRNOT00000057562 |
Myocd
|
myocardin |
| chr17_-_87826421 | 8.96 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chrX_+_111735820 | 8.89 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr2_-_188559882 | 8.88 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr18_+_81694808 | 8.88 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr11_-_782954 | 8.83 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr9_-_105693357 | 8.78 |
ENSRNOT00000066968
|
Nudt12
|
nudix hydrolase 12 |
| chrM_-_14061 | 8.74 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr4_+_94696965 | 8.74 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr6_-_106971250 | 8.72 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr19_+_39229754 | 8.70 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr1_+_80141630 | 8.69 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr8_+_49441106 | 8.66 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr4_-_161850875 | 8.66 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr2_-_89310946 | 8.50 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr3_+_53563194 | 8.50 |
ENSRNOT00000048300
|
Xirp2
|
xin actin-binding repeat containing 2 |
| chr18_+_51523758 | 8.48 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr5_-_168734296 | 8.43 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr8_+_122076759 | 8.41 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chrX_+_77065397 | 8.40 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr12_-_35979193 | 8.29 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr9_+_95501778 | 8.23 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr17_+_8489266 | 8.22 |
ENSRNOT00000016252
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_-_65073012 | 8.10 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr5_-_156105867 | 8.07 |
ENSRNOT00000078851
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr14_+_37116492 | 8.07 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr13_-_82005741 | 8.06 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr8_+_117906014 | 8.05 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr8_+_85503224 | 8.01 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr9_-_85243001 | 7.98 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr18_-_24057917 | 7.96 |
ENSRNOT00000023874
|
Rit2
|
Ras-like without CAAX 2 |
| chr16_+_46731403 | 7.93 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr17_-_32158538 | 7.93 |
ENSRNOT00000024141
|
Nqo2
|
NAD(P)H quinone dehydrogenase 2 |
| chr1_-_275882444 | 7.91 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr9_+_10941613 | 7.88 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chrM_+_14136 | 7.87 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr12_-_10335499 | 7.84 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr9_-_63641400 | 7.82 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr13_-_82006005 | 7.80 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr8_-_87419564 | 7.80 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr5_+_22392732 | 7.78 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr16_-_39476025 | 7.75 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr2_+_66940057 | 7.63 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr8_-_63750531 | 7.62 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr2_-_261337163 | 7.60 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chr3_+_69549673 | 7.54 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr4_+_33638709 | 7.43 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr7_-_76294663 | 7.41 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr6_-_1003905 | 7.36 |
ENSRNOT00000082071
ENSRNOT00000042735 |
Fez2
|
fasciculation and elongation protein zeta 2 |
| chr4_+_29978739 | 7.32 |
ENSRNOT00000011756
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr10_-_29450644 | 7.31 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr2_+_206314213 | 7.28 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr2_+_143475323 | 7.22 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr9_+_73418607 | 7.21 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr3_-_138683318 | 7.12 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr16_+_50049828 | 7.09 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr4_-_17594598 | 7.06 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr3_-_14229067 | 7.05 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr18_+_32336102 | 7.04 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr3_+_151335292 | 7.02 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr13_+_44424689 | 6.97 |
ENSRNOT00000005206
|
Acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr16_-_8885797 | 6.95 |
ENSRNOT00000073370
|
RGD1564899
|
similar to chromosome 10 open reading frame 71 |
| chr4_+_180062799 | 6.89 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chrX_+_74200972 | 6.87 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr2_+_72006099 | 6.87 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr2_+_144861455 | 6.80 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr8_-_8524643 | 6.78 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chrX_+_110789269 | 6.75 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_-_104166367 | 6.71 |
ENSRNOT00000092211
|
Csrp3
|
cysteine and glycine rich protein 3 |
| chr2_-_26699333 | 6.70 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr7_-_6971557 | 6.69 |
ENSRNOT00000050153
|
Olr960
|
olfactory receptor 960 |
| chr9_-_4876023 | 6.68 |
ENSRNOT00000065839
|
RGD1559960
|
similar to Sulfotransferase K1 (rSULT1C2) |
| chr17_-_43537293 | 6.66 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr1_+_107344904 | 6.65 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-1_Evx1_Hesx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 45.8 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 9.0 | 35.9 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 8.2 | 24.7 | GO:2001055 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 8.1 | 24.3 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 7.3 | 21.8 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 5.3 | 15.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 5.2 | 15.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 5.0 | 15.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 4.5 | 22.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 4.2 | 16.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 4.0 | 27.9 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 3.9 | 11.8 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 3.8 | 15.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 3.6 | 46.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 3.5 | 10.5 | GO:1990743 | protein sialylation(GO:1990743) |
| 3.5 | 7.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 3.5 | 10.4 | GO:0048378 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) |
| 3.5 | 3.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 3.2 | 12.7 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 3.1 | 33.9 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 3.1 | 30.5 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 3.0 | 9.0 | GO:2000721 | regulation of cell growth by extracellular stimulus(GO:0001560) phenotypic switching(GO:0036166) negative regulation of beta-amyloid clearance(GO:1900222) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 2.9 | 8.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 2.9 | 14.6 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) gamma-aminobutyric acid receptor clustering(GO:0097112) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 2.9 | 8.8 | GO:0019677 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 2.9 | 8.7 | GO:1903774 | multivesicular body assembly(GO:0036258) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 2.8 | 5.6 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 2.7 | 8.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 2.7 | 13.3 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 2.5 | 7.5 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 2.4 | 7.3 | GO:0045819 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 2.4 | 40.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 2.4 | 18.9 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 2.3 | 11.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.2 | 9.0 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 2.2 | 6.7 | GO:1903919 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 2.2 | 10.8 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 2.1 | 25.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 2.1 | 16.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 2.0 | 10.1 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 2.0 | 20.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 2.0 | 55.8 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 2.0 | 15.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 2.0 | 17.7 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 1.9 | 36.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 1.9 | 20.8 | GO:0009642 | response to light intensity(GO:0009642) |
| 1.8 | 7.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.7 | 27.6 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 1.7 | 6.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.7 | 5.1 | GO:0033306 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 1.7 | 21.8 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 1.7 | 3.3 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 1.6 | 6.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.6 | 11.4 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.6 | 20.4 | GO:0007614 | short-term memory(GO:0007614) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.6 | 4.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.6 | 42.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 1.5 | 10.4 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 1.5 | 4.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) |
| 1.5 | 93.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 1.4 | 5.8 | GO:0050955 | thermoception(GO:0050955) |
| 1.4 | 5.8 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.4 | 5.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.4 | 11.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.4 | 4.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.4 | 4.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 1.4 | 5.6 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 1.4 | 13.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.4 | 8.3 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 1.4 | 5.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.3 | 5.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 1.3 | 3.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.2 | 10.0 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 1.2 | 8.7 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.2 | 13.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 1.2 | 3.6 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.2 | 7.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 1.1 | 5.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 1.1 | 5.7 | GO:0071694 | protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 1.1 | 8.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 1.1 | 6.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 1.1 | 12.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 1.1 | 3.4 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 1.1 | 4.4 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 1.1 | 3.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.1 | 1.1 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 1.1 | 8.7 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 1.1 | 15.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.1 | 3.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 1.1 | 4.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 1.0 | 10.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 1.0 | 9.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 1.0 | 4.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 1.0 | 11.0 | GO:0015747 | urate transport(GO:0015747) |
| 1.0 | 20.6 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 1.0 | 2.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.0 | 16.4 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 1.0 | 15.3 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.9 | 0.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.9 | 8.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.9 | 4.6 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.9 | 3.7 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.9 | 2.7 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.9 | 8.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.9 | 3.5 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.8 | 4.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.8 | 1.6 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.8 | 9.7 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.8 | 5.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.8 | 6.4 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.8 | 12.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.7 | 4.5 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.7 | 2.2 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.7 | 22.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.7 | 5.0 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.7 | 4.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.7 | 10.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.7 | 7.6 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.7 | 3.5 | GO:0097688 | glutamate receptor clustering(GO:0097688) |
| 0.7 | 4.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.7 | 3.3 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.7 | 3.9 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.6 | 7.0 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.6 | 25.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.6 | 4.3 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.6 | 1.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.6 | 7.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.6 | 9.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.6 | 7.9 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.6 | 17.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.6 | 5.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.5 | 4.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.5 | 74.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.5 | 10.2 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.5 | 8.0 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.5 | 3.7 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.5 | 4.7 | GO:1904116 | response to vasopressin(GO:1904116) |
| 0.5 | 7.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.5 | 1.6 | GO:0009227 | nucleotide-sugar catabolic process(GO:0009227) |
| 0.5 | 4.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.5 | 13.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.5 | 8.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 1.0 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.5 | 4.5 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.5 | 2.9 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.5 | 9.8 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) regulation of serine-type peptidase activity(GO:1902571) |
| 0.5 | 8.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.5 | 3.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.5 | 8.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.5 | 1.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.5 | 3.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.5 | 1.4 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.5 | 4.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.5 | 4.5 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.4 | 4.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.4 | 8.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.4 | 1.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.4 | 17.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.4 | 4.7 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.4 | 1.7 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.4 | 4.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.4 | 12.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.4 | 1.2 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.4 | 4.4 | GO:1904181 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.4 | 8.4 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 2.0 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.4 | 1.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.4 | 1.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 4.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.4 | 3.4 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.4 | 0.7 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 1.1 | GO:1990168 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.3 | 2.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.3 | 7.7 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.3 | 2.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 4.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.3 | 2.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.3 | 7.1 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.3 | 3.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 0.7 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.3 | 2.6 | GO:0030647 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.3 | 1.6 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 5.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.3 | 4.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.3 | 1.6 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.3 | 2.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 1.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 8.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 3.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 1.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 2.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.3 | 9.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 2.5 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.3 | 2.3 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 2.8 | GO:0051197 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.3 | 5.0 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.3 | 6.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 2.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 1.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 5.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.3 | 2.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.3 | 0.8 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.3 | 2.6 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.3 | 1.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.3 | 0.8 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.0 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.2 | 0.7 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 2.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 8.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.2 | 0.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 0.5 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 0.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 6.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 13.0 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.2 | 4.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.2 | 5.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.2 | 3.0 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 6.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 3.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 0.7 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.2 | 6.0 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 9.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 13.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 15.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.2 | 5.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.2 | 35.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.2 | 2.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 5.4 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 1.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.2 | 0.8 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 4.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 2.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.2 | 0.5 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.2 | 1.8 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 0.2 | 3.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 3.2 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.2 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 2.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.2 | 2.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 204.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 4.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 3.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.7 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 1.9 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 7.8 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 1.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.1 | 0.4 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 2.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.7 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 2.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 14.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.8 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 2.8 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.1 | 0.1 | GO:0010034 | response to acetate(GO:0010034) |
| 0.1 | 5.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 2.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 10.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 1.7 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.1 | 1.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 0.4 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 2.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 16.5 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 1.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.1 | 4.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.9 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 3.9 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 1.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.4 | GO:0036233 | glycine import(GO:0036233) |
| 0.1 | 1.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.2 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 2.6 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 3.9 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.5 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 4.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 5.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 2.9 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.0 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 1.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 3.2 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 1.1 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) ATP transport(GO:0015867) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 1.0 | GO:0097480 | synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 3.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 10.3 | GO:0090534 | calcium ion-transporting ATPase complex(GO:0090534) |
| 7.0 | 62.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 6.7 | 20.0 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 4.2 | 33.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 4.1 | 24.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 3.9 | 77.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 3.8 | 15.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 2.6 | 39.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 2.5 | 10.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 2.0 | 8.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 1.9 | 95.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.8 | 7.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 1.8 | 8.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.6 | 4.8 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 1.5 | 26.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 1.4 | 4.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 1.3 | 8.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.3 | 19.9 | GO:0031045 | dense core granule(GO:0031045) |
| 1.2 | 8.7 | GO:0090543 | Flemming body(GO:0090543) |
| 1.2 | 8.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.2 | 4.7 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 1.2 | 36.9 | GO:0031430 | M band(GO:0031430) |
| 1.2 | 3.5 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 1.1 | 14.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.0 | 19.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.0 | 12.0 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.9 | 10.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.9 | 10.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.9 | 4.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.9 | 2.6 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.8 | 31.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.7 | 4.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.7 | 8.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 23.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.7 | 10.9 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.7 | 11.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 6.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.6 | 13.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 4.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 10.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 23.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.6 | 29.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.6 | 3.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.6 | 8.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.5 | 3.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.5 | 55.8 | GO:0016605 | PML body(GO:0016605) |
| 0.5 | 2.0 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.5 | 1.9 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.4 | 5.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.4 | 12.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 19.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.4 | 4.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 10.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.4 | 1.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 14.7 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.4 | 3.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 30.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.4 | 31.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 6.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.4 | 7.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 5.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 4.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.4 | 4.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 2.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 1.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 26.0 | GO:0005901 | caveola(GO:0005901) |
| 0.3 | 18.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 3.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 4.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 1.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 1.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 4.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 14.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 5.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 5.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 3.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 3.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 1.9 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.2 | 17.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 3.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 3.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.6 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 19.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 4.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.2 | 3.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 2.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 112.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 1.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 51.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.2 | 10.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.2 | 8.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 8.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 10.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 14.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 20.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 7.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 41.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 13.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 6.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 5.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 25.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 9.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.7 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) autolysosome(GO:0044754) |
| 0.1 | 2.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.7 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 6.6 | GO:0098794 | postsynapse(GO:0098794) |
| 0.1 | 0.5 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 20.5 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 63.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 3.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 4.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 5.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 3.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 10.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 3.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.6 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 45.8 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 7.2 | 28.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 7.0 | 27.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 5.3 | 15.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 5.1 | 20.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 4.2 | 4.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 4.1 | 24.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 3.8 | 15.1 | GO:0019002 | GMP binding(GO:0019002) |
| 3.7 | 14.6 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 3.6 | 97.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 3.5 | 10.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 3.4 | 10.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 3.2 | 9.5 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 3.2 | 12.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 3.0 | 15.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 3.0 | 15.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 3.0 | 11.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 2.8 | 75.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 2.7 | 13.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 2.6 | 7.9 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 2.6 | 23.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 2.6 | 5.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 2.4 | 7.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 2.3 | 7.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 2.2 | 8.8 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 2.0 | 26.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.8 | 26.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.7 | 10.4 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 1.7 | 8.7 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.7 | 5.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 1.7 | 6.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.7 | 5.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.7 | 11.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.6 | 6.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 1.6 | 11.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.5 | 7.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 1.5 | 12.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.5 | 30.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.5 | 5.9 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 1.4 | 4.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.4 | 4.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.3 | 10.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.3 | 49.4 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 1.3 | 7.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 1.3 | 5.1 | GO:0019166 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.2 | 19.5 | GO:0005542 | folic acid binding(GO:0005542) |
| 1.2 | 4.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 1.2 | 11.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.2 | 3.5 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 1.1 | 5.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.1 | 33.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 1.1 | 3.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 1.1 | 5.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.1 | 4.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 1.1 | 10.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.1 | 12.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 1.0 | 8.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 1.0 | 8.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.0 | 16.8 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.9 | 4.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.9 | 6.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.9 | 20.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.9 | 8.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.9 | 6.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.9 | 26.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.9 | 4.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.9 | 3.5 | GO:0003973 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) |
| 0.9 | 1.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.8 | 4.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.8 | 2.5 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.8 | 3.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.8 | 2.5 | GO:0031766 | galanin receptor binding(GO:0031763) type 2 galanin receptor binding(GO:0031765) type 3 galanin receptor binding(GO:0031766) |
| 0.8 | 4.9 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.8 | 3.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.8 | 10.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.8 | 10.1 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.8 | 7.6 | GO:0070700 | co-receptor binding(GO:0039706) BMP receptor binding(GO:0070700) |
| 0.7 | 2.8 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.7 | 4.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.7 | 2.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.7 | 3.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.7 | 30.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 4.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.6 | 16.7 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.6 | 1.9 | GO:0030977 | taurine binding(GO:0030977) |
| 0.6 | 41.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.6 | 7.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.6 | 24.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.6 | 4.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 2.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.6 | 10.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 0.5 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.5 | 3.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 5.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 3.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 3.7 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.5 | 2.6 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.5 | 2.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.5 | 4.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.5 | 24.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.5 | 7.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 2.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 7.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 2.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 4.8 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.5 | 4.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 7.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 3.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 1.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.4 | 5.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 5.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 2.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.4 | 11.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.4 | 4.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.4 | 6.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 11.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.4 | 1.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 12.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 2.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 5.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 38.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.4 | 2.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 2.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 26.4 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.3 | 9.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 2.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 1.6 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.3 | 8.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 4.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 3.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 4.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 3.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.3 | 1.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 0.9 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 2.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 4.0 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.3 | 7.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 2.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 6.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 16.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.3 | 6.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 14.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 4.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 4.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 5.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 1.6 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.3 | 25.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.3 | 24.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.3 | 0.8 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.7 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.2 | 1.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 3.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 4.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 13.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.2 | 2.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 2.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 4.2 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.2 | 4.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.2 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 1.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.2 | 0.9 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 7.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 5.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 1.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.2 | 2.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 3.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 80.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 2.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 7.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 3.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 15.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 4.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 1.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 7.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 3.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 42.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 14.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 13.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 3.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 14.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 5.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 4.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 2.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 120.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 3.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.5 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 0.1 | 0.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 2.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 72.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 3.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 6.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 5.4 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 3.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 2.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 2.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 2.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 4.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 3.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 3.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 4.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 4.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.2 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 4.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 25.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.7 | 33.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.6 | 4.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 6.6 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.5 | 37.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.5 | 13.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 9.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.4 | 27.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.4 | 3.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 4.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.4 | 21.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.4 | 5.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.4 | 4.7 | ST ADRENERGIC | Adrenergic Pathway |
| 0.3 | 5.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 14.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 48.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 16.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 5.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 13.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 14.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 13.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.3 | 4.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 5.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 7.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 8.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 9.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 3.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 6.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 4.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 8.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 6.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 6.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 6.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.2 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 5.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 5.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 27.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 8.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 6.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 6.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 11.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 2.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 45.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 2.6 | 23.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.1 | 33.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.5 | 33.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.3 | 34.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.1 | 11.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.0 | 12.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.9 | 12.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.9 | 12.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.8 | 22.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.8 | 12.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.5 | 7.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.5 | 11.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.5 | 9.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.4 | 27.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 11.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 23.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 8.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.4 | 8.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.4 | 3.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 9.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.3 | 4.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 10.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 20.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 11.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.3 | 8.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 5.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 7.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 7.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 15.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.3 | 5.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.2 | 3.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 7.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 9.2 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.2 | 4.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 6.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 4.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 6.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 3.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 9.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 4.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 15.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 27.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 7.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 5.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 14.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 1.6 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 5.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.7 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 2.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |