Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
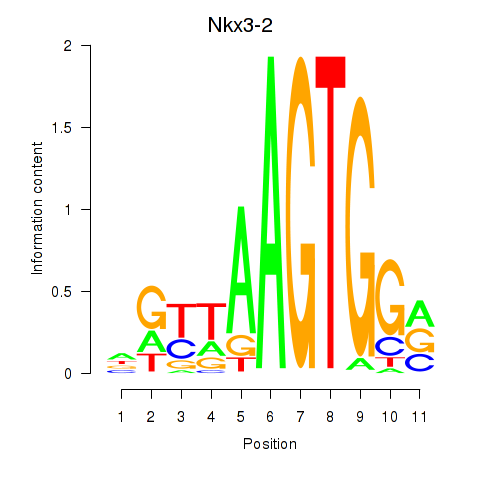
Results for Nkx3-2
Z-value: 1.38
Transcription factors associated with Nkx3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-2
|
ENSRNOG00000060782 | NK3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx3-2 | rn6_v1_chr14_+_73813528_73813528 | -0.39 | 3.7e-13 | Click! |
Activity profile of Nkx3-2 motif
Sorted Z-values of Nkx3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_45561390 | 45.99 |
ENSRNOT00000047820
|
AABR07070043.1
|
|
| chr2_-_200762492 | 35.94 |
ENSRNOT00000056172
|
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr2_-_200762206 | 35.37 |
ENSRNOT00000068511
ENSRNOT00000086835 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr5_+_115649046 | 35.08 |
ENSRNOT00000041328
|
LOC108351137
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr5_-_138222534 | 34.64 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chr1_-_220136470 | 33.92 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chr17_+_32082937 | 32.67 |
ENSRNOT00000074775
|
Mylk4
|
myosin light chain kinase family, member 4 |
| chr4_+_62019970 | 32.20 |
ENSRNOT00000013133
|
LOC100910708
|
aldose reductase-related protein 1-like |
| chr16_+_25773602 | 31.70 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr11_-_72109964 | 29.99 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr5_+_164808323 | 29.02 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr3_+_132052612 | 28.88 |
ENSRNOT00000030148
|
AABR07053879.1
|
|
| chr18_-_63825408 | 28.40 |
ENSRNOT00000043709
|
RGD1562758
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr13_-_111765944 | 28.16 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr16_-_49574314 | 25.68 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr12_+_28381982 | 25.48 |
ENSRNOT00000076101
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 |
| chr8_+_100260049 | 25.40 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr2_+_226825635 | 25.37 |
ENSRNOT00000042173
|
AABR07013200.1
|
|
| chr16_+_16949232 | 24.45 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr10_-_56558487 | 24.43 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr8_-_120446455 | 24.13 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr3_+_55910177 | 23.51 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr5_-_17061837 | 23.39 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr13_-_103080920 | 23.17 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr11_+_70687500 | 22.88 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr5_+_90338795 | 22.53 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr18_+_79406381 | 21.85 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr5_-_17061361 | 21.15 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr20_+_44060731 | 19.92 |
ENSRNOT00000000737
|
Lama4
|
laminin subunit alpha 4 |
| chr2_-_22385676 | 19.85 |
ENSRNOT00000065224
|
Thbs4
|
thrombospondin 4 |
| chr7_-_119158173 | 19.78 |
ENSRNOT00000067483
ENSRNOT00000078528 |
Txn2
|
thioredoxin 2 |
| chr1_-_282251257 | 19.15 |
ENSRNOT00000015186
|
Prdx3
|
peroxiredoxin 3 |
| chr3_+_57770948 | 18.83 |
ENSRNOT00000038429
|
AC107446.2
|
|
| chr12_-_22472044 | 18.73 |
ENSRNOT00000073356
|
Ufsp1
|
UFM1-specific peptidase 1 |
| chr11_-_75144903 | 18.48 |
ENSRNOT00000002329
|
Hrasls
|
HRAS-like suppressor |
| chrX_-_37353156 | 18.25 |
ENSRNOT00000086120
|
AABR07038021.1
|
|
| chrX_-_66602458 | 17.91 |
ENSRNOT00000076510
ENSRNOT00000017429 |
Eda2r
|
ectodysplasin A2 receptor |
| chr2_-_200625434 | 17.87 |
ENSRNOT00000079754
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr18_+_31396922 | 17.70 |
ENSRNOT00000026273
|
RGD735029
|
SEL1 domain containing protein RGD735029 |
| chr8_-_109576353 | 16.77 |
ENSRNOT00000010320
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr1_+_199351628 | 16.67 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr3_+_40038336 | 16.51 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr7_+_116632506 | 15.41 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chrX_+_25016401 | 15.36 |
ENSRNOT00000059270
|
Clcn4
|
chloride voltage-gated channel 4 |
| chr10_-_84698886 | 15.04 |
ENSRNOT00000067542
|
Nfe2l1
|
nuclear factor, erythroid 2-like 1 |
| chr1_-_189960073 | 14.91 |
ENSRNOT00000088482
|
Crym
|
crystallin, mu |
| chr9_-_89195273 | 14.42 |
ENSRNOT00000022281
|
Sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr5_-_25583834 | 13.83 |
ENSRNOT00000067823
|
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr5_+_70441123 | 13.67 |
ENSRNOT00000087517
|
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr2_+_248398917 | 13.35 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr6_+_27589657 | 13.22 |
ENSRNOT00000038649
|
Hadha
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr4_+_129574264 | 13.19 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr4_+_115713004 | 13.17 |
ENSRNOT00000064536
|
Dysf
|
dysferlin |
| chr4_+_115712850 | 12.93 |
ENSRNOT00000085765
|
Dysf
|
dysferlin |
| chr10_-_102289837 | 12.64 |
ENSRNOT00000044922
|
AABR07030729.1
|
|
| chr2_+_166140112 | 12.30 |
ENSRNOT00000074564
|
Ppm1l
|
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr3_-_111469518 | 12.09 |
ENSRNOT00000006874
|
Ndufaf1
|
NADH:ubiquinone oxidoreductase complex assembly factor 1 |
| chr4_+_14039977 | 11.96 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr6_-_55647665 | 11.90 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_13924531 | 11.82 |
ENSRNOT00000001477
|
Slc29a4
|
solute carrier family 29 member 4 |
| chr14_-_37770059 | 11.82 |
ENSRNOT00000029721
|
Slc10a4
|
solute carrier family 10, member 4 |
| chrX_+_109940350 | 11.75 |
ENSRNOT00000093543
ENSRNOT00000057049 |
Nrk
|
Nik related kinase |
| chr2_+_143656793 | 11.73 |
ENSRNOT00000084527
ENSRNOT00000017453 |
Postn
|
periostin |
| chr16_+_81665540 | 10.92 |
ENSRNOT00000026546
ENSRNOT00000092644 ENSRNOT00000073240 |
Grtp1
|
growth hormone regulated TBC protein 1 |
| chr12_+_38377034 | 10.90 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr13_+_90514336 | 10.42 |
ENSRNOT00000088996
ENSRNOT00000085377 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr1_+_80777014 | 10.22 |
ENSRNOT00000079758
|
Gm19345
|
predicted gene, 19345 |
| chr8_+_44136496 | 10.21 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr17_-_90071408 | 9.89 |
ENSRNOT00000071978
|
Pdss1
|
decaprenyl diphosphate synthase subunit 1 |
| chr5_-_164747083 | 9.83 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr10_-_29196563 | 9.64 |
ENSRNOT00000005205
|
Fabp6
|
fatty acid binding protein 6 |
| chr13_+_82355471 | 9.50 |
ENSRNOT00000030677
|
Sele
|
selectin E |
| chr2_+_266315036 | 9.47 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr8_+_62723788 | 9.21 |
ENSRNOT00000010620
|
Sema7a
|
semaphorin 7A (John Milton Hagen blood group) |
| chr13_+_56262190 | 9.18 |
ENSRNOT00000032908
|
AABR07021086.1
|
|
| chr10_-_69537882 | 9.18 |
ENSRNOT00000031360
|
Ccl1
|
C-C motif chemokine ligand 1 |
| chr10_-_89918427 | 9.11 |
ENSRNOT00000084217
|
Dusp3
|
dual specificity phosphatase 3 |
| chr18_+_14756684 | 9.01 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_42121636 | 8.97 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr2_-_171196395 | 8.45 |
ENSRNOT00000013279
|
Bche
|
butyrylcholinesterase |
| chr2_+_151314818 | 8.43 |
ENSRNOT00000019305
|
P2ry1
|
purinergic receptor P2Y1 |
| chr10_-_29196252 | 8.34 |
ENSRNOT00000083910
|
Fabp6
|
fatty acid binding protein 6 |
| chr5_-_166430254 | 8.26 |
ENSRNOT00000048914
|
Nmnat1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr13_-_78885464 | 8.15 |
ENSRNOT00000003828
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr8_-_109418872 | 8.15 |
ENSRNOT00000021657
|
Pccb
|
propionyl-CoA carboxylase beta subunit |
| chrX_+_105500173 | 8.08 |
ENSRNOT00000040476
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr10_+_110138586 | 8.03 |
ENSRNOT00000086096
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr7_-_120672350 | 7.99 |
ENSRNOT00000018126
ENSRNOT00000087800 ENSRNOT00000088424 |
Csnk1e
|
casein kinase 1, epsilon |
| chr10_-_78690857 | 7.96 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_89491654 | 7.93 |
ENSRNOT00000028632
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chr2_+_187512164 | 7.78 |
ENSRNOT00000051394
|
Mef2d
|
myocyte enhancer factor 2D |
| chr19_+_57047830 | 7.76 |
ENSRNOT00000080860
|
Galnt2
|
polypeptide N-acetylgalactosaminyltransferase 2 |
| chr2_+_66940057 | 7.73 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr19_-_1074333 | 7.65 |
ENSRNOT00000017983
ENSRNOT00000086995 |
Cdh5
|
cadherin 5 |
| chr6_+_108167716 | 7.59 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr13_+_82355886 | 7.55 |
ENSRNOT00000076757
|
Sele
|
selectin E |
| chr1_-_755645 | 7.50 |
ENSRNOT00000073546
|
Vom2r6
|
vomeronasal 2 receptor, 6 |
| chr8_-_83693472 | 7.43 |
ENSRNOT00000015947
|
Fam83b
|
family with sequence similarity 83, member B |
| chr9_+_62291405 | 7.42 |
ENSRNOT00000051463
|
Plcl1
|
phospholipase C-like 1 |
| chr20_-_4390436 | 7.42 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr10_-_36419926 | 7.30 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr12_-_23624212 | 7.28 |
ENSRNOT00000064405
ENSRNOT00000001943 |
Rasa4
|
RAS p21 protein activator 4 |
| chr2_-_189903219 | 7.18 |
ENSRNOT00000017000
|
S100a1
|
S100 calcium binding protein A1 |
| chr2_-_147693082 | 7.15 |
ENSRNOT00000022507
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr17_-_21577631 | 7.07 |
ENSRNOT00000066371
|
Tmem14c
|
transmembrane protein 14C |
| chr12_+_40018937 | 7.00 |
ENSRNOT00000001697
|
Cux2
|
cut-like homeobox 2 |
| chr10_+_88326080 | 6.96 |
ENSRNOT00000076475
|
Fkbp10
|
FK506 binding protein 10 |
| chr8_-_90664554 | 6.76 |
ENSRNOT00000039917
ENSRNOT00000074896 ENSRNOT00000072225 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr4_+_109467272 | 6.75 |
ENSRNOT00000008212
|
Reg3b
|
regenerating family member 3 beta |
| chrX_-_74968405 | 6.73 |
ENSRNOT00000035653
|
RGD1561931
|
similar to KIAA2022 protein |
| chr9_+_73528681 | 6.62 |
ENSRNOT00000084340
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr5_+_140870140 | 6.38 |
ENSRNOT00000074347
|
Hpcal4
|
hippocalcin-like 4 |
| chr17_-_84249243 | 6.12 |
ENSRNOT00000082549
|
Nebl
|
nebulette |
| chr1_-_64030175 | 6.11 |
ENSRNOT00000089950
|
Tsen34l1
|
tRNA splicing endonuclease subunit 34-like 1 |
| chr1_+_240908483 | 6.07 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr7_+_38897278 | 6.07 |
ENSRNOT00000006450
|
Epyc
|
epiphycan |
| chr11_-_81717521 | 6.05 |
ENSRNOT00000058422
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr11_-_87081950 | 6.00 |
ENSRNOT00000002574
|
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr3_-_175426395 | 5.62 |
ENSRNOT00000082686
|
Psma7
|
proteasome subunit alpha 7 |
| chr16_-_21089508 | 5.61 |
ENSRNOT00000072565
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr10_+_88326337 | 5.56 |
ENSRNOT00000022029
ENSRNOT00000076461 |
Fkbp10
|
FK506 binding protein 10 |
| chr10_+_85646633 | 5.53 |
ENSRNOT00000086608
|
Psmb3
|
proteasome subunit beta 3 |
| chr1_+_699413 | 5.44 |
ENSRNOT00000072360
|
AABR07000089.1
|
|
| chr19_+_45938915 | 5.40 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr1_-_67065797 | 5.38 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr1_-_172943853 | 5.34 |
ENSRNOT00000047040
|
Olr278
|
olfactory receptor 278 |
| chr10_-_27862868 | 5.30 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr3_+_114253637 | 5.30 |
ENSRNOT00000046843
|
Duox1
|
dual oxidase 1 |
| chr9_+_62291809 | 5.24 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr18_+_30574627 | 5.13 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr7_+_123168811 | 5.09 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr14_-_115352562 | 5.06 |
ENSRNOT00000050321
|
Erlec1
|
endoplasmic reticulum lectin 1 |
| chr10_-_34333305 | 4.96 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr8_-_107819050 | 4.93 |
ENSRNOT00000019536
|
Armc8
|
armadillo repeat containing 8 |
| chr3_+_20993205 | 4.85 |
ENSRNOT00000010465
|
Olr419
|
olfactory receptor 419 |
| chr10_-_88533829 | 4.83 |
ENSRNOT00000080521
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr11_+_75434197 | 4.82 |
ENSRNOT00000032569
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr3_+_147358858 | 4.82 |
ENSRNOT00000012951
|
Rspo4
|
R-spondin 4 |
| chr3_+_72460889 | 4.77 |
ENSRNOT00000012209
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr19_+_56243408 | 4.74 |
ENSRNOT00000000281
|
Def8
|
differentially expressed in FDCP 8 homolog |
| chr9_-_86103158 | 4.61 |
ENSRNOT00000021528
|
Cul3
|
cullin 3 |
| chr8_-_117820413 | 4.61 |
ENSRNOT00000075819
|
Tma7
|
translation machinery associated 7 homolog |
| chr1_-_63457134 | 4.57 |
ENSRNOT00000083436
|
AABR07071874.1
|
|
| chr15_+_23792931 | 4.54 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chrX_+_158835811 | 4.49 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr1_-_168611670 | 4.45 |
ENSRNOT00000021273
|
Olr106
|
olfactory receptor 106 |
| chr9_-_10710648 | 4.40 |
ENSRNOT00000089374
|
Kdm4b
|
lysine demethylase 4B |
| chr10_-_103816287 | 4.30 |
ENSRNOT00000004477
|
Grin2c
|
glutamate ionotropic receptor NMDA type subunit 2C |
| chr14_+_109533792 | 4.27 |
ENSRNOT00000067690
|
LOC108348154
|
ankyrin repeat domain-containing protein 29-like |
| chr7_+_59326518 | 4.10 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr8_+_21305250 | 4.10 |
ENSRNOT00000050206
|
Olr1193
|
olfactory receptor 1193 |
| chr12_+_12374790 | 4.08 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr3_-_105470475 | 4.03 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr1_-_71135764 | 4.03 |
ENSRNOT00000064949
|
Smim17
|
small integral membrane protein 17 |
| chr17_-_33951484 | 4.00 |
ENSRNOT00000023957
|
Foxc1
|
forkhead box C1 |
| chr3_+_2512492 | 3.96 |
ENSRNOT00000074026
|
AABR07051241.1
|
|
| chr13_-_89565813 | 3.92 |
ENSRNOT00000004347
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr10_-_1744647 | 3.86 |
ENSRNOT00000081886
|
AABR07028997.1
|
|
| chrX_-_123662350 | 3.83 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr10_-_74298599 | 3.81 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr3_+_20878701 | 3.79 |
ENSRNOT00000046877
|
Olr413
|
olfactory receptor 413 |
| chr1_+_226742795 | 3.78 |
ENSRNOT00000084270
|
NEWGENE_1309258
|
VPS37C, ESCRT-I subunit |
| chr12_-_52396066 | 3.78 |
ENSRNOT00000056712
|
Lrcol1
|
leucine rich colipase-like 1 |
| chr8_-_21341431 | 3.75 |
ENSRNOT00000044249
|
Olr1192
|
olfactory receptor 1192 |
| chr13_-_72063347 | 3.73 |
ENSRNOT00000090544
ENSRNOT00000003869 |
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr14_+_216098 | 3.71 |
ENSRNOT00000033032
|
Vom2r65
|
vomeronasal 2 receptor, 65 |
| chr8_+_117648745 | 3.66 |
ENSRNOT00000027762
|
Slc26a6
|
solute carrier family 26 member 6 |
| chr20_-_4391402 | 3.62 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr5_+_79367663 | 3.55 |
ENSRNOT00000011508
|
Atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr10_-_34361212 | 3.52 |
ENSRNOT00000072557
|
Olr1385
|
olfactory receptor 1385 |
| chrX_+_73390903 | 3.48 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr7_+_98432024 | 3.48 |
ENSRNOT00000091077
|
Fer1l6
|
fer-1-like family member 6 |
| chr1_+_113034227 | 3.46 |
ENSRNOT00000081831
ENSRNOT00000077877 ENSRNOT00000077594 |
Gabrb3
|
gamma-aminobutyric acid type A receptor beta 3 subunit |
| chr1_+_67025240 | 3.43 |
ENSRNOT00000051024
|
Vom1r47
|
vomeronasal 1 receptor 47 |
| chr2_+_58462949 | 3.43 |
ENSRNOT00000080618
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr7_-_24667301 | 3.39 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chr16_-_64745207 | 3.39 |
ENSRNOT00000032467
|
Tti2
|
TELO2 interacting protein 2 |
| chr10_+_94207336 | 3.37 |
ENSRNOT00000010816
|
Kcnh6
|
potassium voltage-gated channel subfamily H member 6 |
| chr18_+_30509393 | 3.29 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr1_-_261446570 | 3.24 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr3_+_16241573 | 3.20 |
ENSRNOT00000082831
|
Olr407
|
olfactory receptor 407 |
| chr10_-_88533171 | 3.13 |
ENSRNOT00000024321
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr6_+_55085313 | 3.12 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr1_-_67206713 | 3.10 |
ENSRNOT00000048195
|
Vom1r43
|
vomeronasal 1 receptor 43 |
| chr8_-_20173107 | 3.08 |
ENSRNOT00000045215
|
Olr1163
|
olfactory receptor 1163 |
| chr3_+_79729739 | 3.07 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr1_-_196884302 | 3.07 |
ENSRNOT00000089464
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr10_+_111770168 | 3.03 |
ENSRNOT00000074598
|
Vom2r65
|
vomeronasal 2 receptor, 65 |
| chr3_-_76289390 | 3.01 |
ENSRNOT00000051968
|
Olr609
|
olfactory receptor 609 |
| chr3_+_118317761 | 2.99 |
ENSRNOT00000012700
|
Fgf7
|
fibroblast growth factor 7 |
| chr10_+_112056994 | 2.96 |
ENSRNOT00000073032
|
Vom2r65
|
vomeronasal 2 receptor, 65 |
| chr15_+_42522108 | 2.95 |
ENSRNOT00000021621
|
Ccdc25
|
coiled-coil domain containing 25 |
| chr8_-_18762922 | 2.89 |
ENSRNOT00000008423
|
Olr1122
|
olfactory receptor 1122 |
| chr10_-_44471915 | 2.83 |
ENSRNOT00000089901
|
Olr1440
|
olfactory receptor 1440 |
| chr6_-_67084234 | 2.83 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr9_+_23596964 | 2.79 |
ENSRNOT00000064279
|
LOC108351994
|
exocrine gland-secreted peptide 1-like |
| chr10_-_66020682 | 2.79 |
ENSRNOT00000011019
|
Fam58b
|
family with sequence similarity 58, member B |
| chr19_-_55490426 | 2.79 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr19_-_10653800 | 2.72 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr9_+_70450444 | 2.71 |
ENSRNOT00000002872
|
Fastkd2
|
FAST kinase domains 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.8 | 44.5 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 11.9 | 71.3 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 11.7 | 35.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 11.3 | 33.9 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 11.2 | 33.6 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 9.7 | 29.0 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 6.1 | 6.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 5.6 | 16.8 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 5.1 | 15.4 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 4.0 | 19.8 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 3.8 | 19.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 3.3 | 9.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 3.2 | 9.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.9 | 8.7 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 2.8 | 8.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 2.7 | 8.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 2.6 | 13.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 2.4 | 12.0 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 2.3 | 38.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 2.2 | 13.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 2.2 | 10.9 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 2.0 | 6.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 2.0 | 26.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 2.0 | 11.8 | GO:0060723 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 1.9 | 7.8 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 1.8 | 5.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 1.8 | 5.3 | GO:0042335 | cuticle development(GO:0042335) |
| 1.7 | 10.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.7 | 6.8 | GO:1903576 | response to L-arginine(GO:1903576) |
| 1.7 | 8.4 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 1.7 | 23.5 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 1.6 | 3.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.5 | 6.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.4 | 4.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.4 | 11.0 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 1.4 | 14.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.3 | 6.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 1.3 | 4.0 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 1.3 | 10.4 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 1.3 | 2.6 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 1.3 | 7.6 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.2 | 19.9 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 1.2 | 3.7 | GO:0046724 | oxalic acid secretion(GO:0046724) |
| 1.2 | 16.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 1.2 | 4.6 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 1.2 | 9.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 1.0 | 12.7 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 1.0 | 13.4 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.9 | 24.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.9 | 13.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.9 | 7.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.9 | 8.0 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.9 | 8.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.8 | 2.3 | GO:0048749 | compound eye development(GO:0048749) |
| 0.8 | 2.3 | GO:0046864 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.7 | 2.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.7 | 3.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.7 | 2.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.7 | 8.0 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.6 | 9.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.6 | 7.8 | GO:0007512 | adult heart development(GO:0007512) |
| 0.6 | 7.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.6 | 11.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.6 | 17.0 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.6 | 2.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.6 | 4.4 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.5 | 1.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.5 | 9.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.5 | 4.8 | GO:0035878 | nail development(GO:0035878) |
| 0.5 | 3.7 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.5 | 18.0 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.5 | 7.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.5 | 3.0 | GO:0060665 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.5 | 5.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.5 | 25.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 2.7 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.4 | 2.8 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.4 | 9.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.4 | 24.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.4 | 11.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 14.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.3 | 2.5 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.3 | 15.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.3 | 6.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.3 | 12.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 7.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.2 | 7.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 0.9 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.2 | 1.2 | GO:2000504 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 17.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 4.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 17.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.2 | 3.1 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 15.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 7.9 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.2 | 1.4 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 4.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 1.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.0 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.1 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.2 | 1.7 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.1 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 7.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 0.6 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.1 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.8 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 8.1 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.1 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.4 | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 14.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 5.1 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 3.8 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 4.0 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.9 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.1 | 0.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.5 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 81.2 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 2.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 1.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 13.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 10.1 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.6 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 1.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 2.6 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 0.2 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 3.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 3.8 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 2.8 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.1 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.4 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 44.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 4.4 | 35.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 3.6 | 21.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 3.3 | 13.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 3.3 | 16.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 2.7 | 26.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.9 | 24.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.8 | 23.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.5 | 19.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 1.3 | 6.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.3 | 3.8 | GO:0032173 | septin collar(GO:0032173) |
| 1.2 | 6.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.1 | 10.9 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.1 | 7.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 1.0 | 29.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.9 | 15.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.7 | 4.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.6 | 17.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.6 | 5.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.6 | 97.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.5 | 17.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.5 | 19.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.5 | 26.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.4 | 7.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 3.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.4 | 8.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 9.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 9.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 1.9 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 15.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 9.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 4.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 4.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 17.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 6.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 3.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 3.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.2 | 4.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 10.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 4.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 30.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 4.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 7.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 12.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 81.5 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 0.9 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 4.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.6 | GO:0071541 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 77.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 32.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 4.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 5.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 7.4 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.8 | 71.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 8.8 | 35.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 7.4 | 44.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 4.9 | 19.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 4.8 | 19.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 4.6 | 13.8 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 4.5 | 17.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 4.4 | 13.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 3.3 | 13.3 | GO:0019002 | GMP binding(GO:0019002) |
| 3.3 | 9.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 2.8 | 8.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 2.8 | 8.3 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 2.7 | 24.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 2.6 | 15.4 | GO:0035473 | lipase binding(GO:0035473) |
| 2.5 | 19.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 2.4 | 12.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 2.2 | 24.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 2.1 | 8.4 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 2.0 | 18.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 2.0 | 11.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.6 | 4.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.6 | 11.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.5 | 4.6 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.5 | 6.1 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 1.5 | 10.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 1.4 | 29.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.4 | 9.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.3 | 10.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.2 | 14.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.2 | 6.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 1.2 | 3.7 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 1.2 | 17.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.1 | 8.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 1.0 | 8.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 1.0 | 8.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.0 | 12.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.9 | 7.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.9 | 12.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.8 | 15.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.7 | 3.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 8.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 45.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.7 | 13.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.6 | 3.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.6 | 26.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 9.1 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.5 | 1.5 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.5 | 10.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 4.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 7.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 17.0 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.4 | 2.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.4 | 4.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 3.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 6.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 15.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 9.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 11.9 | GO:0008009 | chemokine activity(GO:0008009) CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 2.3 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.2 | 36.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 4.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 5.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.6 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 8.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.2 | 1.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 1.9 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 2.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.9 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 4.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 2.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 4.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 8.2 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 9.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 3.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 12.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 12.9 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 14.4 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.1 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 9.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 6.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 12.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.8 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 4.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 7.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 5.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 2.1 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 8.9 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 9.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 65.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 5.1 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 22.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 4.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.8 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 4.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.1 | GO:0015459 | calcium-activated potassium channel activity(GO:0015269) potassium channel regulator activity(GO:0015459) |
| 0.0 | 6.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 14.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 3.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 71.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.8 | 24.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.7 | 7.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.6 | 19.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 21.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.4 | 8.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 7.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 10.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 7.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 11.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 21.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 11.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 6.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 15.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 19.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 69.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 1.5 | 21.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.5 | 18.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.4 | 24.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.3 | 36.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.0 | 21.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 1.0 | 49.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.7 | 8.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.7 | 9.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.6 | 8.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 10.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 4.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 8.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 9.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 13.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.4 | 8.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 12.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 15.8 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.3 | 10.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 21.0 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.3 | 7.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.3 | 4.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 13.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 4.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 36.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 4.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 3.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.2 | 2.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 2.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 8.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 19.8 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 5.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 3.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 4.4 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 2.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 4.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |