Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
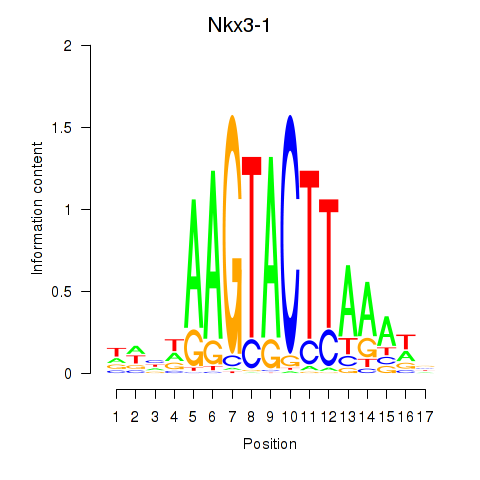
Results for Nkx3-1
Z-value: 0.64
Transcription factors associated with Nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-1
|
ENSRNOG00000015477 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx3-1 | rn6_v1_chr15_+_51065316_51065316 | 0.10 | 6.5e-02 | Click! |
Activity profile of Nkx3-1 motif
Sorted Z-values of Nkx3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_120446455 | 21.10 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr11_+_54619129 | 13.20 |
ENSRNOT00000059924
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr15_+_31642169 | 13.15 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chr4_+_70572942 | 13.11 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr15_+_31417147 | 12.31 |
ENSRNOT00000092182
|
AABR07017824.2
|
|
| chr15_-_29761117 | 12.17 |
ENSRNOT00000075194
|
AABR07017658.2
|
|
| chr19_-_10681145 | 11.68 |
ENSRNOT00000022167
|
Ccl22
|
C-C motif chemokine ligand 22 |
| chr15_+_31097291 | 11.65 |
ENSRNOT00000075456
|
AABR07017789.1
|
|
| chr1_-_104202591 | 11.31 |
ENSRNOT00000035512
|
E2f8
|
E2F transcription factor 8 |
| chr15_+_30570483 | 11.22 |
ENSRNOT00000071740
|
AABR07017745.1
|
|
| chr10_-_31419235 | 9.89 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr15_+_31010891 | 9.50 |
ENSRNOT00000088134
|
AABR07017781.1
|
|
| chr2_-_203413124 | 9.48 |
ENSRNOT00000077898
ENSRNOT00000056140 |
Cd101
|
CD101 molecule |
| chr2_-_132551235 | 9.41 |
ENSRNOT00000058250
|
LOC100365525
|
rCG65904-like |
| chr9_+_60900213 | 8.95 |
ENSRNOT00000017859
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr2_+_127625683 | 8.77 |
ENSRNOT00000015474
|
Hspa4l
|
heat shock protein 4-like |
| chr15_-_30019055 | 8.65 |
ENSRNOT00000073984
|
AABR07017681.1
|
|
| chr4_-_128266082 | 8.29 |
ENSRNOT00000039549
|
Nup50
|
nucleoporin 50 |
| chrX_+_70461718 | 8.26 |
ENSRNOT00000078233
ENSRNOT00000003789 |
Kif4a
|
kinesin family member 4A |
| chr15_-_29246222 | 7.85 |
ENSRNOT00000081806
|
AABR07017617.1
|
|
| chr8_-_13906355 | 7.70 |
ENSRNOT00000029634
|
Cep295
|
centrosomal protein 295 |
| chr13_-_95348913 | 7.70 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr15_-_30323833 | 7.66 |
ENSRNOT00000071631
|
AABR07017714.1
|
|
| chr1_+_253221812 | 7.13 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr15_-_45524582 | 6.98 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr4_-_69163881 | 6.86 |
ENSRNOT00000048779
|
Tryx5
|
trypsin X5 |
| chr4_+_52199416 | 6.85 |
ENSRNOT00000009537
|
Spam1
|
sperm adhesion molecule 1 |
| chr1_-_205630073 | 6.62 |
ENSRNOT00000037064
|
Tex36
|
testis expressed 36 |
| chr3_+_80468154 | 6.22 |
ENSRNOT00000021837
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr3_+_148790979 | 6.16 |
ENSRNOT00000085757
|
Kif3b
|
kinesin family member 3B |
| chr1_-_205669760 | 6.09 |
ENSRNOT00000086703
|
Tex36
|
testis expressed 36 |
| chr15_-_88670349 | 5.90 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr2_-_165591110 | 5.85 |
ENSRNOT00000091140
|
Ift80
|
intraflagellar transport 80 |
| chr3_+_15379109 | 5.83 |
ENSRNOT00000036495
|
Morn5
|
MORN repeat containing 5 |
| chr6_-_25507073 | 5.81 |
ENSRNOT00000061176
|
Plb1
|
phospholipase B1 |
| chr14_-_3446611 | 5.73 |
ENSRNOT00000080901
|
Brdt
|
bromodomain testis associated |
| chr1_+_81579736 | 5.67 |
ENSRNOT00000071339
|
LOC100910611
|
CD177 antigen-like |
| chr12_+_37878653 | 5.65 |
ENSRNOT00000091084
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr3_+_150055749 | 5.53 |
ENSRNOT00000055335
|
Actl10
|
actin-like 10 |
| chr1_+_156552328 | 5.50 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr5_+_16845631 | 5.40 |
ENSRNOT00000047889
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_-_176596591 | 5.36 |
ENSRNOT00000086365
|
Recql
|
RecQ like helicase |
| chr15_+_32012543 | 5.34 |
ENSRNOT00000078174
|
AABR07017868.5
|
|
| chr14_+_81858737 | 5.33 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chrX_-_40086870 | 5.27 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr7_+_40217991 | 5.26 |
ENSRNOT00000085684
|
Cep290
|
centrosomal protein 290 |
| chr19_-_37819789 | 5.22 |
ENSRNOT00000036702
|
Cenpt
|
centromere protein T |
| chrX_+_29504349 | 5.08 |
ENSRNOT00000005981
|
Tceanc
|
transcription elongation factor A N-terminal and central domain containing |
| chr1_+_106405002 | 5.08 |
ENSRNOT00000074027
|
LOC103690168
|
probable isocitrate dehydrogenase [NAD] gamma 2, mitochondrial |
| chr19_+_56220755 | 5.06 |
ENSRNOT00000023452
|
Tubb3
|
tubulin, beta 3 class III |
| chr1_+_82816984 | 5.05 |
ENSRNOT00000074588
|
LOC100910611
|
CD177 antigen-like |
| chr4_-_6046477 | 5.00 |
ENSRNOT00000073925
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr18_-_74478341 | 4.99 |
ENSRNOT00000074572
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr15_-_33285779 | 4.99 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chr19_+_38162515 | 4.98 |
ENSRNOT00000027117
|
Slc7a6
|
solute carrier family 7 member 6 |
| chr10_-_17721233 | 4.95 |
ENSRNOT00000031337
|
Smim23
|
small integral membrane protein 23 |
| chr1_+_82934134 | 4.94 |
ENSRNOT00000072914
|
LOC100910611
|
CD177 antigen-like |
| chr15_+_47455690 | 4.90 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr11_+_42936028 | 4.86 |
ENSRNOT00000074653
|
AABR07033887.1
|
|
| chr4_+_22898527 | 4.79 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr1_+_252497452 | 4.74 |
ENSRNOT00000075598
|
Stambpl1
|
STAM binding protein-like 1 |
| chr1_-_189199376 | 4.72 |
ENSRNOT00000021027
|
Umod
|
uromodulin |
| chr10_+_109950988 | 4.68 |
ENSRNOT00000071293
|
LOC688321
|
similar to Lung carbonyl reductase [NADPH] (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27) |
| chr15_-_55209342 | 4.57 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr4_+_160020472 | 4.52 |
ENSRNOT00000078802
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr7_+_27620458 | 4.47 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr10_+_72147816 | 4.47 |
ENSRNOT00000090085
|
LOC102553386
|
SUMO-conjugating enzyme UBC9-like |
| chr14_+_3204390 | 4.46 |
ENSRNOT00000088078
|
Glmn
|
glomulin, FKBP associated protein |
| chr2_+_202200797 | 4.41 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr4_-_6062641 | 4.38 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr16_+_34795971 | 4.27 |
ENSRNOT00000043510
|
LOC100912321
|
myeloid-associated differentiation marker-like |
| chr8_-_78397123 | 4.21 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr11_+_53081025 | 4.21 |
ENSRNOT00000002700
|
Bbx
|
BBX, HMG-box containing |
| chr3_-_112371664 | 4.19 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chr6_-_27460038 | 4.14 |
ENSRNOT00000036815
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr9_-_27761365 | 4.14 |
ENSRNOT00000018552
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr1_+_101410019 | 4.09 |
ENSRNOT00000042039
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr1_-_277867630 | 3.93 |
ENSRNOT00000054679
ENSRNOT00000091038 ENSRNOT00000079869 |
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_58149061 | 3.84 |
ENSRNOT00000005157
|
Tph2
|
tryptophan hydroxylase 2 |
| chr2_+_143895982 | 3.81 |
ENSRNOT00000080026
|
Supt20h
|
SPT20 homolog, SAGA complex component |
| chr2_-_54963448 | 3.78 |
ENSRNOT00000017886
|
Ptger4
|
prostaglandin E receptor 4 |
| chr16_+_56247659 | 3.73 |
ENSRNOT00000017452
|
Tusc3
|
tumor suppressor candidate 3 |
| chr8_+_5606592 | 3.71 |
ENSRNOT00000011727
|
Mmp12
|
matrix metallopeptidase 12 |
| chr1_-_277939039 | 3.71 |
ENSRNOT00000079579
ENSRNOT00000086726 |
Ablim1
|
actin-binding LIM protein 1 |
| chr13_-_109770199 | 3.69 |
ENSRNOT00000043672
|
Fam71a
|
family with sequence similarity 71, member A |
| chr14_+_100135271 | 3.67 |
ENSRNOT00000032965
|
Fbxo48
|
F-box protein 48 |
| chr10_+_90985653 | 3.63 |
ENSRNOT00000093364
|
Ccdc103
|
coiled-coil domain containing 103 |
| chr2_+_123791335 | 3.63 |
ENSRNOT00000077096
|
Adad1
|
adenosine deaminase domain containing 1 |
| chr20_-_27578244 | 3.62 |
ENSRNOT00000000708
|
Fam26d
|
family with sequence similarity 26, member D |
| chr15_+_31895223 | 3.60 |
ENSRNOT00000092202
|
AABR07017855.1
|
|
| chr7_+_127964752 | 3.60 |
ENSRNOT00000038168
|
RGD1560617
|
hypothetical gene supported by NM_053561; AF062594 |
| chr3_+_54734015 | 3.56 |
ENSRNOT00000034341
|
LOC685184
|
similar to protein expressed in prostate, ovary, testis, and placenta 8 isoform 2 |
| chr10_+_11879418 | 3.54 |
ENSRNOT00000068577
|
RGD1561796
|
RGD1561796 |
| chr9_-_27761733 | 3.51 |
ENSRNOT00000040034
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr4_+_159403501 | 3.48 |
ENSRNOT00000086525
|
Akap3
|
A-kinase anchoring protein 3 |
| chr9_-_15700235 | 3.42 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr3_-_161294125 | 3.41 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chr13_+_90723092 | 3.41 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr13_-_82295123 | 3.39 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr20_-_399933 | 3.37 |
ENSRNOT00000000953
|
Olr1671
|
olfactory receptor 1671 |
| chrX_-_158261717 | 3.36 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr10_-_7029136 | 3.35 |
ENSRNOT00000091942
|
AC129395.1
|
|
| chr17_-_57526461 | 3.35 |
ENSRNOT00000072723
|
LOC103690053
|
ankyrin repeat domain-containing protein 26-like |
| chr9_-_81772851 | 3.34 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr1_+_101249522 | 3.25 |
ENSRNOT00000033882
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr2_-_137331140 | 3.17 |
ENSRNOT00000048546
|
LOC365791
|
similar to T-cell activation Rho GTPase-activating protein isoform b |
| chr8_-_50603591 | 3.13 |
ENSRNOT00000077748
|
LOC500990
|
similar to RIKEN cDNA 4931429L15 |
| chr14_+_7630482 | 3.10 |
ENSRNOT00000091581
|
LOC498330
|
similar to hypothetical protein MGC26744 |
| chr1_-_82901917 | 3.05 |
ENSRNOT00000035648
|
LOC102553892
|
CD177 antigen-like |
| chr2_+_43271092 | 3.04 |
ENSRNOT00000017571
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr10_-_70181708 | 3.00 |
ENSRNOT00000076598
|
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr3_+_146695344 | 2.99 |
ENSRNOT00000010955
|
Gins1
|
GINS complex subunit 1 |
| chr3_-_112385528 | 2.99 |
ENSRNOT00000049756
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr18_+_70996044 | 2.96 |
ENSRNOT00000024950
ENSRNOT00000066336 |
Dym
|
dymeclin |
| chr5_+_147714163 | 2.95 |
ENSRNOT00000012663
|
Marcksl1
|
MARCKS-like 1 |
| chr17_+_85500113 | 2.90 |
ENSRNOT00000091937
|
AABR07028768.1
|
|
| chr6_+_27887797 | 2.89 |
ENSRNOT00000015827
|
Asxl2
|
additional sex combs like 2, transcriptional regulator |
| chr2_-_39007976 | 2.86 |
ENSRNOT00000064292
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr19_-_14945302 | 2.78 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr5_+_135413275 | 2.71 |
ENSRNOT00000012263
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chrX_-_124162058 | 2.67 |
ENSRNOT00000071436
|
LOC367975
|
similar to B-cell translocation gene 1 |
| chr10_+_65161152 | 2.65 |
ENSRNOT00000011608
|
Cryba1
|
crystallin, beta A1 |
| chrX_-_38468360 | 2.54 |
ENSRNOT00000045285
|
Map7d2
|
MAP7 domain containing 2 |
| chr6_-_55698757 | 2.54 |
ENSRNOT00000085146
|
Lrrc72
|
leucine rich repeat containing 72 |
| chr1_-_20962526 | 2.54 |
ENSRNOT00000061332
ENSRNOT00000017322 ENSRNOT00000017412 ENSRNOT00000079688 ENSRNOT00000017417 |
Epb41l2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr9_+_24066303 | 2.53 |
ENSRNOT00000018163
|
Crisp3
|
cysteine-rich secretory protein 3 |
| chr5_-_133067245 | 2.51 |
ENSRNOT00000033163
|
Skint10
|
selection and upkeep of intraepithelial T cells 10 |
| chr1_-_7480825 | 2.50 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr11_-_66759402 | 2.49 |
ENSRNOT00000003326
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr8_+_117600749 | 2.49 |
ENSRNOT00000047482
|
Nckipsd
|
NCK interacting protein with SH3 domain |
| chrX_-_82699487 | 2.49 |
ENSRNOT00000081625
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr13_-_105684374 | 2.47 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr1_-_81559685 | 2.43 |
ENSRNOT00000075282
|
LOC100909620
|
CD177 antigen-like |
| chr12_+_12859661 | 2.42 |
ENSRNOT00000001404
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr13_-_15395617 | 2.40 |
ENSRNOT00000046060
|
LOC304725
|
similar to contactin associated protein-like 5 isoform 1 |
| chr6_+_102477036 | 2.38 |
ENSRNOT00000087132
|
Rad51b
|
RAD51 paralog B |
| chr1_-_99632845 | 2.37 |
ENSRNOT00000049123
|
LOC103691107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 pseudogene |
| chr14_+_48726045 | 2.34 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr20_-_1818800 | 2.33 |
ENSRNOT00000000990
|
RT1-M3-1
|
RT1 class Ib, locus M3, gene 1 |
| chr12_+_19303387 | 2.32 |
ENSRNOT00000001820
|
Cops6
|
COP9 signalosome subunit 6 |
| chr13_+_89524329 | 2.32 |
ENSRNOT00000004279
|
Mpz
|
myelin protein zero |
| chr4_-_117531480 | 2.29 |
ENSRNOT00000021120
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr2_+_86914989 | 2.29 |
ENSRNOT00000085164
ENSRNOT00000081966 |
LOC100360380
|
zinc finger protein 457-like |
| chr9_+_71230108 | 2.27 |
ENSRNOT00000018326
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr15_-_70399924 | 2.22 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr6_+_26390689 | 2.21 |
ENSRNOT00000079762
ENSRNOT00000091528 |
Ift172
|
intraflagellar transport 172 |
| chr7_-_116504853 | 2.20 |
ENSRNOT00000056557
|
RGD1565410
|
similar to Ly6-C antigen gene |
| chr18_-_36513317 | 2.18 |
ENSRNOT00000025453
|
Plac8l1
|
PLAC8-like 1 |
| chr17_-_10439691 | 2.15 |
ENSRNOT00000024151
|
Cdhr2
|
cadherin-related family member 2 |
| chr2_-_198942732 | 2.10 |
ENSRNOT00000000109
ENSRNOT00000073913 |
Cd160
|
CD160 molecule |
| chr15_+_2526368 | 2.08 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chr3_-_21609853 | 2.04 |
ENSRNOT00000012160
|
Pdcl
|
phosducin-like |
| chr1_+_255564409 | 2.04 |
ENSRNOT00000081786
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr2_-_49170025 | 2.04 |
ENSRNOT00000060595
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_+_30863217 | 1.97 |
ENSRNOT00000015421
|
Rnf146
|
ring finger protein 146 |
| chr1_+_81372650 | 1.91 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr7_+_74350479 | 1.90 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr3_+_101899068 | 1.90 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr19_-_56799445 | 1.89 |
ENSRNOT00000024491
|
RGD1563991
|
similar to TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr8_-_39201588 | 1.88 |
ENSRNOT00000011226
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr7_-_139542461 | 1.86 |
ENSRNOT00000089425
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr3_-_114728410 | 1.82 |
ENSRNOT00000000181
|
Gatm
|
glycine amidinotransferase |
| chr4_-_129430251 | 1.81 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr12_+_4546287 | 1.81 |
ENSRNOT00000001416
|
Higd2al1
|
HIG1 hypoxia inducible domain family, member 2A-like 1 |
| chr15_+_31051414 | 1.80 |
ENSRNOT00000090250
|
AABR07017783.4
|
|
| chr1_+_13595295 | 1.80 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr20_+_34895059 | 1.79 |
ENSRNOT00000084420
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr12_-_47438931 | 1.78 |
ENSRNOT00000050782
|
RGD1311899
|
similar to RIKEN cDNA 2210016L21 gene |
| chr7_-_68549763 | 1.78 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr9_-_65957101 | 1.77 |
ENSRNOT00000014094
|
Mpp4
|
membrane palmitoylated protein 4 |
| chr18_+_87415531 | 1.76 |
ENSRNOT00000072475
|
AABR07032888.1
|
|
| chr5_+_137546860 | 1.76 |
ENSRNOT00000074431
|
Olr858
|
olfactory receptor 858 |
| chr11_+_87224421 | 1.75 |
ENSRNOT00000000308
|
Dgcr14
|
DiGeorge syndrome critical region gene 14 |
| chr14_-_6284145 | 1.74 |
ENSRNOT00000044065
|
Zfp951
|
zinc finger protein 951 |
| chr14_-_6358720 | 1.74 |
ENSRNOT00000048419
|
Zfp951
|
zinc finger protein 951 |
| chr7_+_2645196 | 1.74 |
ENSRNOT00000083899
|
Timeless
|
timeless circadian clock |
| chr1_-_165891627 | 1.73 |
ENSRNOT00000038408
|
Relt
|
RELT tumor necrosis factor receptor |
| chr13_+_71110064 | 1.64 |
ENSRNOT00000088329
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr5_+_8876044 | 1.60 |
ENSRNOT00000008877
|
Cops5
|
COP9 signalosome subunit 5 |
| chr10_+_34414854 | 1.59 |
ENSRNOT00000074980
|
LOC684471
|
similar to olfactory receptor 1394 |
| chrX_-_21945305 | 1.58 |
ENSRNOT00000065084
|
Samt2
|
spermatogenesis associated multipass transmembrane protein 2 |
| chr8_-_48727154 | 1.56 |
ENSRNOT00000088471
|
Trappc4
|
trafficking protein particle complex 4 |
| chr9_-_82336806 | 1.50 |
ENSRNOT00000024667
|
Slc23a3
|
solute carrier family 23, member 3 |
| chr18_+_15738809 | 1.50 |
ENSRNOT00000075444
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr8_-_48726963 | 1.44 |
ENSRNOT00000016145
|
Trappc4
|
trafficking protein particle complex 4 |
| chr9_+_100885051 | 1.43 |
ENSRNOT00000082987
|
AC109427.1
|
|
| chr1_-_67065797 | 1.42 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr18_+_3163214 | 1.39 |
ENSRNOT00000017291
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr1_-_66639205 | 1.36 |
ENSRNOT00000082190
|
LOC100909481
|
eukaryotic translation initiation factor 3 subunit E-like |
| chr4_-_118487633 | 1.36 |
ENSRNOT00000068093
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr12_-_30304036 | 1.35 |
ENSRNOT00000001218
|
Nupr2
|
nuclear protein 2, transcriptional regulator |
| chr3_-_111266542 | 1.33 |
ENSRNOT00000031672
|
Ino80
|
INO80 complex subunit |
| chr2_-_231877307 | 1.31 |
ENSRNOT00000073860
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr6_+_125876109 | 1.26 |
ENSRNOT00000091300
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr2_-_144808245 | 1.24 |
ENSRNOT00000087849
|
LOC102552324
|
uncharacterized LOC102552324 |
| chr17_-_42226377 | 1.22 |
ENSRNOT00000024536
|
RGD1307443
|
similar to mKIAA0319 protein |
| chr6_+_83083740 | 1.18 |
ENSRNOT00000007600
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr8_+_17928358 | 1.16 |
ENSRNOT00000029911
|
Mbd3l2
|
methyl-CpG binding domain protein 3-like 2 |
| chr5_+_139783951 | 1.15 |
ENSRNOT00000081333
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr2_+_189714754 | 1.15 |
ENSRNOT00000086285
|
Gatad2b
|
GATA zinc finger domain containing 2B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.9 | 5.7 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 1.9 | 11.3 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.6 | 13.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.5 | 4.6 | GO:1903943 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 1.3 | 3.8 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 1.2 | 9.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.2 | 4.7 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 1.2 | 8.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 1.1 | 3.4 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 1.1 | 4.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.0 | 6.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 3.7 | GO:1902741 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.8 | 5.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.8 | 2.3 | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.8 | 2.3 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.6 | 5.8 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.6 | 3.8 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.6 | 3.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.6 | 2.5 | GO:0042531 | regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.6 | 5.8 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.6 | 1.7 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 0.5 | 6.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.5 | 2.0 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.5 | 3.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.5 | 5.5 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.5 | 2.9 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.5 | 1.8 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.4 | 5.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.4 | 5.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.4 | 2.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.4 | 5.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.4 | 2.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 3.6 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.4 | 3.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 1.1 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.3 | 4.5 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.3 | 5.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 0.9 | GO:0046671 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) cellular response to fluoride(GO:1902618) |
| 0.3 | 21.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.3 | 2.5 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.3 | 7.7 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 3.4 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.3 | 3.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.3 | 3.0 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.3 | 2.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.3 | 1.1 | GO:0071651 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.3 | 1.9 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.3 | 2.7 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 | 3.7 | GO:0015693 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.3 | 1.9 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 | 9.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.2 | 11.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 0.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.2 | 1.4 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 1.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.2 | 1.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.0 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 4.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 0.6 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.2 | 2.6 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 2.2 | GO:0061525 | hindgut development(GO:0061525) |
| 0.2 | 0.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 0.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 5.1 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 0.5 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 1.6 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 9.5 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.1 | 11.1 | GO:0046068 | cGMP metabolic process(GO:0046068) |
| 0.1 | 6.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 4.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.7 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 5.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 5.0 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 0.6 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.1 | 2.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 8.8 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 1.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 8.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 4.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 7.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 2.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.0 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 1.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 4.9 | GO:1902117 | positive regulation of organelle assembly(GO:1902117) |
| 0.1 | 3.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.3 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 1.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:1904430 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:0097240 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.8 | GO:2000650 | negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 2.0 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.5 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 2.3 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 2.1 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.2 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 5.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.6 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 2.0 | GO:0090263 | protein autoubiquitination(GO:0051865) positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.6 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 | 1.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.5 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 4.0 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 6.1 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 2.4 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 2.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.2 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.4 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 1.8 | GO:0007601 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 1.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 4.7 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.1 | 2.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.1 | 5.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.0 | 3.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.9 | 4.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.8 | 2.3 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.8 | 4.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.8 | 2.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.8 | 3.0 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.7 | 13.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.7 | 9.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.6 | 11.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.5 | 2.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.5 | 7.1 | GO:1990023 | mitotic spindle pole(GO:0097431) mitotic spindle midzone(GO:1990023) |
| 0.4 | 4.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.4 | 2.2 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 5.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 5.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 3.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 7.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 2.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 3.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 5.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 8.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 3.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 5.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 3.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 1.4 | GO:0033290 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.2 | 1.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 8.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 13.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 8.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 5.9 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.1 | 0.9 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 7.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 6.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 7.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 4.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.5 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 4.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 7.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 6.4 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 2.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 1.5 | 4.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 1.3 | 5.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.2 | 5.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.0 | 15.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.9 | 4.6 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.9 | 5.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.9 | 3.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.8 | 2.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.7 | 6.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.5 | 5.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.5 | 3.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.5 | 3.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.5 | 2.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.5 | 5.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 5.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 4.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.4 | 5.0 | GO:0070097 | alpha-catenin binding(GO:0045294) delta-catenin binding(GO:0070097) |
| 0.4 | 1.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.4 | 1.9 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.4 | 4.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 1.0 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.3 | 5.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.3 | 3.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.3 | 4.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 9.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 11.7 | GO:0008009 | chemokine activity(GO:0008009) CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 7.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 4.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 8.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 5.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 2.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 4.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 0.5 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.6 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 2.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.2 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 5.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 7.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 6.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 5.0 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 7.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.8 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 3.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 9.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 6.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 1.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 10.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 8.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 3.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 6.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 3.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 12.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 2.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.5 | 3.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.5 | 13.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.5 | 14.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 4.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 7.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 8.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.3 | 5.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 3.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 3.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 4.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 4.8 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 11.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 13.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 5.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 4.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 7.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 5.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.3 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 1.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.4 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 3.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 5.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |