Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
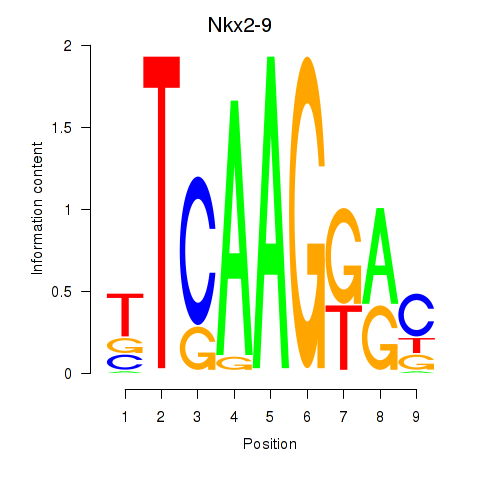
Results for Nkx2-9
Z-value: 1.06
Transcription factors associated with Nkx2-9
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-8 | rn6_v1_chr6_-_77508585_77508585 | 0.32 | 2.9e-09 | Click! |
Activity profile of Nkx2-9 motif
Sorted Z-values of Nkx2-9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_107952530 | 46.58 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr6_-_138662365 | 40.53 |
ENSRNOT00000066209
ENSRNOT00000084892 |
Ighm
|
immunoglobulin heavy constant mu |
| chr6_-_142903440 | 31.74 |
ENSRNOT00000075707
|
AABR07065823.2
|
|
| chr10_-_31493419 | 28.27 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr6_-_140880070 | 24.73 |
ENSRNOT00000073779
|
LOC691828
|
uncharacterized LOC691828 |
| chr6_-_140102325 | 24.25 |
ENSRNOT00000072238
|
AABR07065750.2
|
|
| chr5_-_157199441 | 24.05 |
ENSRNOT00000022559
|
Pla2g2f
|
phospholipase A2, group IIF |
| chr14_+_48726045 | 21.41 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_-_105390580 | 21.23 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr7_+_28654733 | 21.02 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr6_+_139405966 | 20.74 |
ENSRNOT00000088974
|
AABR07065693.3
|
|
| chr6_-_142353308 | 20.00 |
ENSRNOT00000066416
|
AABR07065814.2
|
|
| chr2_-_241472694 | 19.83 |
ENSRNOT00000051498
|
Bank1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr6_-_143702033 | 19.31 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr6_-_143131118 | 18.74 |
ENSRNOT00000074930
|
AABR07065834.1
|
|
| chr15_+_61826937 | 16.53 |
ENSRNOT00000084005
ENSRNOT00000079417 |
Elf1
|
E74-like factor 1 |
| chr14_+_108826831 | 16.13 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr4_-_131694755 | 15.97 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr9_+_81427730 | 15.32 |
ENSRNOT00000019109
ENSRNOT00000081711 |
Cxcr2
|
C-X-C motif chemokine receptor 2 |
| chr1_+_279798187 | 14.41 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr6_-_138736203 | 13.77 |
ENSRNOT00000052021
|
LOC100360169
|
rCG21044-like |
| chr4_+_145489869 | 13.12 |
ENSRNOT00000082618
|
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr20_-_4132604 | 12.97 |
ENSRNOT00000077630
ENSRNOT00000048332 |
RT1-Da
|
RT1 class II, locus Da |
| chr6_-_140642221 | 12.63 |
ENSRNOT00000081996
|
AABR07065772.2
|
|
| chr15_-_29369504 | 12.51 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chr1_-_174047572 | 12.04 |
ENSRNOT00000019597
|
Stk33
|
serine/threonine kinase 33 |
| chrX_+_120624518 | 11.97 |
ENSRNOT00000007967
|
Slc6a14
|
solute carrier family 6 member 14 |
| chr9_-_93404883 | 11.87 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr17_-_54808483 | 11.82 |
ENSRNOT00000075683
|
AABR07028049.1
|
|
| chr8_+_29453643 | 11.70 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr2_+_251983339 | 11.69 |
ENSRNOT00000020230
|
Mcoln3
|
mucolipin 3 |
| chr7_-_12346475 | 11.36 |
ENSRNOT00000060708
|
Mum1
|
melanoma associated antigen (mutated) 1 |
| chr14_+_39964588 | 11.35 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr7_+_141355994 | 11.35 |
ENSRNOT00000081195
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr16_+_22979444 | 11.32 |
ENSRNOT00000017822
|
Csgalnact1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr6_+_52663112 | 11.29 |
ENSRNOT00000013842
|
Atxn7l1
|
ataxin 7-like 1 |
| chr7_-_55604403 | 10.75 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr11_+_81358592 | 10.69 |
ENSRNOT00000002487
|
Rfc4
|
replication factor C subunit 4 |
| chr5_-_12526962 | 10.58 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr9_-_29337922 | 10.05 |
ENSRNOT00000018896
|
RGD1309049
|
similar to RIKEN cDNA 4933415F23 |
| chr1_-_84812486 | 9.69 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr3_+_172719432 | 9.36 |
ENSRNOT00000033863
|
Zfp831
|
zinc finger protein 831 |
| chr1_-_233140237 | 9.34 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr1_+_256786124 | 9.17 |
ENSRNOT00000034563
|
Ffar4
|
free fatty acid receptor 4 |
| chr10_+_45893018 | 9.16 |
ENSRNOT00000004280
ENSRNOT00000086710 |
Nlrp3
|
NLR family, pyrin domain containing 3 |
| chr2_-_60657712 | 9.13 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr1_-_7801438 | 9.01 |
ENSRNOT00000022273
|
AABR07000276.1
|
|
| chr5_-_58455819 | 8.81 |
ENSRNOT00000078082
|
Fancg
|
Fanconi anemia, complementation group G |
| chr1_+_156552328 | 8.59 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr7_-_68549763 | 8.50 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr5_-_137112927 | 7.84 |
ENSRNOT00000078302
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F |
| chr5_+_139963002 | 7.60 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr6_-_108660063 | 7.32 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr19_-_14945302 | 7.32 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr4_+_145413230 | 7.22 |
ENSRNOT00000056508
|
Il17re
|
interleukin 17 receptor E |
| chr5_+_122019301 | 7.17 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr15_+_80040842 | 6.91 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chrX_-_70428364 | 6.79 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr1_+_220423426 | 6.69 |
ENSRNOT00000072647
|
Brms1
|
breast cancer metastasis-suppressor 1 |
| chr6_-_145777767 | 6.35 |
ENSRNOT00000046799
|
Rpl32
|
ribosomal protein L32 |
| chr13_+_51972974 | 6.34 |
ENSRNOT00000008164
|
Arl8a
|
ADP-ribosylation factor like GTPase 8A |
| chr5_-_153184940 | 5.86 |
ENSRNOT00000079536
|
Tmem57
|
transmembrane protein 57 |
| chr3_-_46051096 | 5.53 |
ENSRNOT00000081302
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr8_-_61917125 | 5.36 |
ENSRNOT00000085049
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr9_+_69953440 | 5.31 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr9_+_81644355 | 5.28 |
ENSRNOT00000071700
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr1_+_161401527 | 5.22 |
ENSRNOT00000015181
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr13_+_44812567 | 5.17 |
ENSRNOT00000005372
|
R3hdm1
|
R3H domain containing 1 |
| chr2_-_188559882 | 5.03 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr18_+_27190974 | 4.66 |
ENSRNOT00000037690
|
Pkd2l2
|
polycystin 2 like 2, transient receptor potential cation channel |
| chr11_+_60072727 | 4.65 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr10_-_15228235 | 4.49 |
ENSRNOT00000027121
|
Wdr90
|
WD repeat domain 90 |
| chr1_+_79831534 | 4.45 |
ENSRNOT00000057965
|
Nova2
|
NOVA alternative splicing regulator 2 |
| chr18_+_30010918 | 4.23 |
ENSRNOT00000084132
|
Pcdha4
|
protocadherin alpha 4 |
| chr10_-_87407634 | 4.17 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr5_-_73494030 | 4.04 |
ENSRNOT00000022291
|
Actl7b
|
actin-like 7b |
| chr17_-_43675934 | 3.92 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr20_+_41184287 | 3.77 |
ENSRNOT00000081117
|
Col10a1
|
collagen type X alpha 1 chain |
| chr11_+_57505005 | 3.77 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr10_-_97647822 | 3.68 |
ENSRNOT00000065781
|
AC106648.1
|
|
| chr2_-_23260965 | 3.49 |
ENSRNOT00000046772
|
LOC103691423
|
60S ribosomal protein L36 pseudogene |
| chr14_+_109533792 | 3.45 |
ENSRNOT00000067690
|
LOC108348154
|
ankyrin repeat domain-containing protein 29-like |
| chr17_+_47721977 | 3.43 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr10_+_70134729 | 3.40 |
ENSRNOT00000076739
|
Lig3
|
DNA ligase 3 |
| chr18_+_27576129 | 3.23 |
ENSRNOT00000070930
|
Kdm3b
|
lysine demethylase 3B |
| chrX_-_15467875 | 3.16 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr19_-_41010122 | 3.04 |
ENSRNOT00000088770
|
Vac14
|
Vac14, PIKFYVE complex component |
| chr18_-_25997555 | 2.90 |
ENSRNOT00000027755
|
Stard4
|
StAR-related lipid transfer domain containing 4 |
| chr10_+_15241590 | 2.89 |
ENSRNOT00000037372
ENSRNOT00000037381 |
Mettl26
|
methyltransferase like 26 |
| chr5_+_50381244 | 2.88 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr6_-_143537030 | 2.76 |
ENSRNOT00000071876
|
LOC100360610
|
rCG64220-like |
| chr11_-_67037115 | 2.62 |
ENSRNOT00000003137
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chrX_-_64908682 | 2.55 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr3_+_161018511 | 2.51 |
ENSRNOT00000019804
ENSRNOT00000039664 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chrX_-_106116886 | 2.47 |
ENSRNOT00000052009
|
Tcp11x2
|
t-complex 11 family, X-linked 2 |
| chr14_-_44375804 | 2.43 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr5_-_127273656 | 2.32 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr15_+_28696378 | 2.28 |
ENSRNOT00000017476
|
Tox4
|
TOX high mobility group box family member 4 |
| chr9_-_15375365 | 2.27 |
ENSRNOT00000018550
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr15_-_58171049 | 2.13 |
ENSRNOT00000001371
|
Gpalpp1
|
GPALPP motifs containing 1 |
| chr15_+_58170967 | 2.09 |
ENSRNOT00000001366
|
Nufip1
|
NUFIP1, FMR1 interacting protein 1 |
| chr15_+_28695912 | 1.96 |
ENSRNOT00000081830
|
Tox4
|
TOX high mobility group box family member 4 |
| chr17_+_54280851 | 1.65 |
ENSRNOT00000024022
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr1_-_90961717 | 1.58 |
ENSRNOT00000071180
|
LOC108348106
|
alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 |
| chr18_-_52017734 | 1.56 |
ENSRNOT00000081020
|
March3
|
membrane associated ring-CH-type finger 3 |
| chr7_+_35773928 | 1.45 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr7_+_142106465 | 1.29 |
ENSRNOT00000042786
ENSRNOT00000088681 |
Letmd1
|
LETM1 domain containing 1 |
| chrX_-_152367861 | 1.12 |
ENSRNOT00000089414
|
LOC103690371
|
melanoma-associated antigen 10-like |
| chr6_+_84293402 | 1.12 |
ENSRNOT00000072657
|
LOC108351365
|
uncharacterized LOC108351365 |
| chr15_-_28696122 | 0.95 |
ENSRNOT00000060467
|
Rab2b
|
RAB2B, member RAS oncogene family |
| chrX_+_128493614 | 0.93 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chr5_-_35831712 | 0.91 |
ENSRNOT00000010126
|
Prdm13
|
PR/SET domain 13 |
| chr12_-_45801842 | 0.89 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr6_-_98666007 | 0.84 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr9_+_99720856 | 0.82 |
ENSRNOT00000047487
|
Olr1346
|
olfactory receptor 1346 |
| chr6_-_93562314 | 0.79 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr8_+_85951191 | 0.74 |
ENSRNOT00000037665
|
Cd109
|
CD109 molecule |
| chr7_+_120923274 | 0.71 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr15_-_29672591 | 0.68 |
ENSRNOT00000060298
|
LOC100911282
|
uncharacterized LOC100911282 |
| chr6_-_141866756 | 0.66 |
ENSRNOT00000068561
|
AABR07065804.1
|
|
| chr12_-_11144754 | 0.53 |
ENSRNOT00000084637
|
Zfp655
|
zinc finger protein 655 |
| chr9_+_66058047 | 0.33 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr5_-_147823232 | 0.29 |
ENSRNOT00000074345
|
Iqcc
|
IQ motif containing C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 46.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 5.4 | 16.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 5.3 | 16.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 5.3 | 21.2 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) cellular response to molecule of fungal origin(GO:0071226) |
| 4.2 | 21.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 3.8 | 11.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 3.1 | 15.3 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 2.6 | 28.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 2.3 | 9.2 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 2.2 | 13.0 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.9 | 13.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.9 | 9.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.7 | 5.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 1.7 | 8.5 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.7 | 19.8 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 1.4 | 11.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.3 | 7.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 1.2 | 7.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.2 | 10.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.1 | 3.4 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.8 | 6.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.8 | 8.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.7 | 6.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.7 | 2.9 | GO:0070859 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.7 | 24.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.6 | 2.3 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 1.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.3 | 11.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 9.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.3 | 5.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.3 | 11.9 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.3 | 2.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 3.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.2 | 9.2 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.2 | 2.9 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.2 | 8.9 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.7 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 8.8 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.1 | 3.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 3.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 6.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 14.4 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.1 | 12.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 3.2 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 3.0 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 7.3 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 2.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 12.0 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 5.3 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.7 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 4.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.6 | GO:0090277 | positive regulation of peptide hormone secretion(GO:0090277) |
| 0.0 | 14.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 11.4 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 4.4 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 4.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 12.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.5 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.9 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 1.8 | 10.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 1.8 | 5.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.3 | 9.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 1.3 | 36.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.1 | 3.4 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 1.0 | 5.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.9 | 11.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.8 | 13.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.6 | 8.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.5 | 8.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 2.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 3.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.4 | 2.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 7.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 4.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 6.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 44.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 11.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 6.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 11.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 19.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 12.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 11.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 8.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 15.7 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 2.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 24.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 9.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 6.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 21.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 5.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.3 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 3.8 | 11.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 2.3 | 6.8 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 2.0 | 7.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.7 | 8.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.1 | 10.7 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.9 | 7.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.9 | 2.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.9 | 8.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.8 | 42.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.8 | 14.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.7 | 24.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.7 | 13.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.7 | 9.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.7 | 11.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.7 | 13.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 21.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.5 | 28.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.4 | 16.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.4 | 12.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.4 | 7.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 3.4 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 9.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.3 | 9.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.2 | 3.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 11.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 6.7 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.2 | 8.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 11.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 16.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 53.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 1.6 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 2.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 6.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 4.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 2.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 11.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 14.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 8.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.9 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 5.5 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 6.3 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 49.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.6 | 32.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.6 | 15.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.5 | 21.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 16.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 9.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 14.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 24.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 1.4 | 46.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 1.3 | 13.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.0 | 28.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.8 | 9.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 10.7 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.6 | 8.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 21.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.6 | 6.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 12.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 9.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.5 | 8.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.5 | 20.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.5 | 11.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 15.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 3.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 7.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 29.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 3.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 7.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 23.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 11.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |