Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
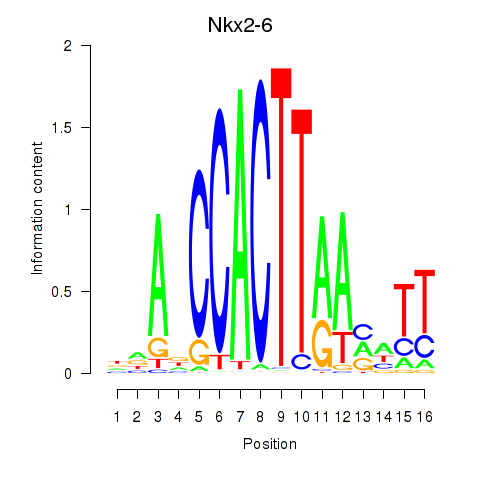
Results for Nkx2-6
Z-value: 0.22
Transcription factors associated with Nkx2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-6
|
ENSRNOG00000021910 | NK2 homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-6 | rn6_v1_chr15_+_51034620_51034620 | -0.62 | 1.8e-35 | Click! |
Activity profile of Nkx2-6 motif
Sorted Z-values of Nkx2-6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_282251257 | 6.57 |
ENSRNOT00000015186
|
Prdx3
|
peroxiredoxin 3 |
| chr2_+_84645084 | 5.34 |
ENSRNOT00000015448
|
Cmbl
|
carboxymethylenebutenolidase homolog |
| chr16_-_56900052 | 4.74 |
ENSRNOT00000017339
|
Msr1
|
macrophage scavenger receptor 1 |
| chr17_-_43537293 | 4.30 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr9_+_62291405 | 3.54 |
ENSRNOT00000051463
|
Plcl1
|
phospholipase C-like 1 |
| chrX_-_157286936 | 2.94 |
ENSRNOT00000078100
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr17_-_81187739 | 2.58 |
ENSRNOT00000063911
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr9_+_3896337 | 1.65 |
ENSRNOT00000079166
|
LOC103693189
|
protein tyrosine phosphatase type IVA 1 |
| chr3_+_140024043 | 1.51 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr4_-_180234804 | 1.35 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr9_+_51009116 | 1.15 |
ENSRNOT00000039313
|
Mettl21cl1
|
methyltransferase like 21C-like 1 |
| chr5_-_115222075 | 1.06 |
ENSRNOT00000058163
ENSRNOT00000012503 |
LOC100912642
|
cytochrome P450 2J3-like |
| chr2_-_26870421 | 0.92 |
ENSRNOT00000088791
|
AABR07007744.2
|
|
| chr1_+_185427736 | 0.83 |
ENSRNOT00000064706
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr4_-_155421998 | 0.83 |
ENSRNOT00000041552
|
Gdf3
|
growth differentiation factor 3 |
| chr7_+_107406962 | 0.77 |
ENSRNOT00000093254
|
Phf20l1
|
PHD finger protein 20-like protein 1 |
| chrX_+_78042859 | 0.76 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr1_-_189460844 | 0.63 |
ENSRNOT00000020077
|
Thumpd1
|
THUMP domain containing 1 |
| chr13_+_98231326 | 0.57 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_203768729 | 0.55 |
ENSRNOT00000018895
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr3_+_15856289 | 0.53 |
ENSRNOT00000089297
|
Olr399
|
olfactory receptor 399 |
| chr17_+_66579319 | 0.48 |
ENSRNOT00000071829
|
Heatr1
|
HEAT repeat containing 1 |
| chr1_-_252922486 | 0.48 |
ENSRNOT00000076802
|
Ifit1bl
|
interferon-induced protein with tetratricopeptide repeats 1B-like |
| chr3_+_16117228 | 0.44 |
ENSRNOT00000049590
|
Olr404
|
olfactory receptor 404 |
| chr10_+_49538588 | 0.37 |
ENSRNOT00000081415
|
Pmp22
|
peripheral myelin protein 22 |
| chr3_-_74014795 | 0.32 |
ENSRNOT00000073426
|
Olr516
|
olfactory receptor 516 |
| chr3_+_20878701 | 0.14 |
ENSRNOT00000046877
|
Olr413
|
olfactory receptor 413 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.9 | 2.6 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.4 | 4.7 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.4 | 4.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.4 | 2.9 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 3.5 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 0.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.3 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 | 0.8 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.5 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.9 | 2.6 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.4 | 4.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 3.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 4.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 1.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 0.8 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 2.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0019840 | isoprenoid binding(GO:0019840) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |