Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
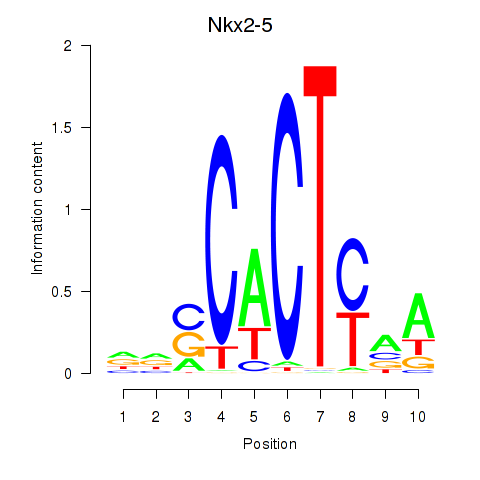
Results for Nkx2-5
Z-value: 0.70
Transcription factors associated with Nkx2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-5
|
ENSRNOG00000020747 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-5 | rn6_v1_chr10_+_16635989_16635989 | 0.56 | 1.7e-27 | Click! |
Activity profile of Nkx2-5 motif
Sorted Z-values of Nkx2-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_164808323 | 41.95 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr16_+_18716019 | 41.27 |
ENSRNOT00000047870
|
Sftpa1
|
surfactant protein A1 |
| chr4_+_100166863 | 32.75 |
ENSRNOT00000014505
|
Sftpb
|
surfactant protein B |
| chr7_+_60099120 | 31.81 |
ENSRNOT00000007338
|
LOC100911101
|
leucine-rich repeat-containing protein 10-like |
| chr7_+_60087429 | 27.85 |
ENSRNOT00000073117
|
Lrrc10
|
leucine-rich repeat-containing 10 |
| chr15_-_14510828 | 11.75 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr1_+_177569618 | 11.36 |
ENSRNOT00000090042
|
Tead1
|
TEA domain transcription factor 1 |
| chr13_-_82005741 | 10.86 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr10_+_40247436 | 10.00 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr10_-_43512287 | 9.11 |
ENSRNOT00000036481
|
Faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr18_+_56071478 | 8.32 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr16_-_64806050 | 6.87 |
ENSRNOT00000015725
|
Dusp26
|
dual specificity phosphatase 26 |
| chr7_-_23594133 | 6.83 |
ENSRNOT00000005746
|
Timp3
|
TIMP metallopeptidase inhibitor 3 |
| chrX_+_37329779 | 6.00 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr17_-_55709740 | 5.89 |
ENSRNOT00000033359
|
RGD1562037
|
similar to OTTHUMP00000046255 |
| chr10_-_44147035 | 5.82 |
ENSRNOT00000041113
|
Olr1424
|
olfactory receptor 1424 |
| chr15_+_23777856 | 5.62 |
ENSRNOT00000060847
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr7_-_114690642 | 5.56 |
ENSRNOT00000078097
|
AABR07058406.1
|
|
| chr7_-_119797098 | 4.98 |
ENSRNOT00000009994
|
Rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr13_-_107886476 | 4.97 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr14_-_114047527 | 4.72 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr10_+_49472460 | 4.66 |
ENSRNOT00000038276
|
Tekt3
|
tektin 3 |
| chr1_-_141470380 | 4.66 |
ENSRNOT00000065759
|
Plin1
|
perilipin 1 |
| chr14_-_34561696 | 4.44 |
ENSRNOT00000059763
|
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr2_-_198458041 | 3.98 |
ENSRNOT00000082450
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr2_-_185444897 | 3.65 |
ENSRNOT00000016329
|
Rps3a
|
ribosomal protein S3a |
| chr10_+_104523996 | 3.32 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr1_+_197813963 | 3.22 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chr12_-_19159901 | 2.90 |
ENSRNOT00000045401
|
Rpl31l4
|
ribosomal protein L31-like 4 |
| chr5_-_154438361 | 2.87 |
ENSRNOT00000085003
|
AC141344.2
|
|
| chr8_-_43480777 | 2.83 |
ENSRNOT00000060074
|
LOC688473
|
similar to 40S ribosomal protein S2 |
| chr16_+_49185560 | 2.81 |
ENSRNOT00000014360
|
Helt
|
helt bHLH transcription factor |
| chr10_+_14828597 | 2.63 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr6_+_28515025 | 2.33 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr14_-_85088523 | 2.33 |
ENSRNOT00000010690
|
Nf2
|
neurofibromin 2 |
| chr2_-_23260965 | 2.24 |
ENSRNOT00000046772
|
LOC103691423
|
60S ribosomal protein L36 pseudogene |
| chr12_-_13462038 | 2.21 |
ENSRNOT00000043110
|
LOC100360439
|
ribosomal protein L36-like |
| chr5_-_78393143 | 2.10 |
ENSRNOT00000043610
|
LOC100910017
|
60S ribosomal protein L31-like |
| chr20_-_4390436 | 1.99 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr1_-_89258935 | 1.98 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr3_+_20965751 | 1.93 |
ENSRNOT00000010450
|
Olr417
|
olfactory receptor 417 |
| chr17_+_36956385 | 1.92 |
ENSRNOT00000024854
|
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr5_-_83648044 | 1.88 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr14_+_82769642 | 1.87 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr9_-_60923706 | 1.78 |
ENSRNOT00000047517
|
AABR07067749.1
|
|
| chr4_+_121760773 | 1.75 |
ENSRNOT00000087069
|
Vom1r92
|
vomeronasal 1 receptor 92 |
| chr3_-_102589260 | 1.74 |
ENSRNOT00000091642
|
AC121203.1
|
|
| chr10_-_44157330 | 1.60 |
ENSRNOT00000047242
|
Olr1425
|
olfactory receptor 1425 |
| chr7_-_143473697 | 1.60 |
ENSRNOT00000055336
ENSRNOT00000083027 |
Krt77
|
keratin 77 |
| chr11_-_61530567 | 1.53 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_+_33440191 | 1.51 |
ENSRNOT00000034632
ENSRNOT00000092907 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr1_+_86362432 | 1.45 |
ENSRNOT00000052300
|
Vom1r2
|
vomeronasal 1 receptor 2 |
| chr9_+_44567206 | 1.41 |
ENSRNOT00000088544
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr5_-_155709215 | 1.29 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr4_+_121912352 | 1.26 |
ENSRNOT00000086882
|
Vom1r98
|
vomeronasal 1 receptor 98 |
| chr10_+_53595854 | 1.22 |
ENSRNOT00000044116
|
Sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr3_+_102841615 | 1.21 |
ENSRNOT00000075178
|
Olr770
|
olfactory receptor 770 |
| chr17_-_36438752 | 1.18 |
ENSRNOT00000048997
|
RGD1561310
|
similar to ribosomal protein L37 |
| chr10_-_44095775 | 1.14 |
ENSRNOT00000047892
|
Olr1422
|
olfactory receptor 1422 |
| chr1_-_230649938 | 1.12 |
ENSRNOT00000017615
|
Olr377
|
olfactory receptor 377 |
| chr20_-_4391402 | 1.12 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr1_-_228558593 | 1.09 |
ENSRNOT00000028606
|
Olr326
|
olfactory receptor 326 |
| chr8_+_3638921 | 1.05 |
ENSRNOT00000075829
|
Vom1r24
|
vomeronasal 1 receptor 24 |
| chr13_-_91638661 | 0.96 |
ENSRNOT00000045429
|
Olr1579
|
olfactory receptor 1579 |
| chr9_-_99365192 | 0.94 |
ENSRNOT00000047012
|
RGD1564730
|
similar to ribosomal protein L36 |
| chr15_+_31178339 | 0.90 |
ENSRNOT00000089064
|
AABR07017798.2
|
|
| chrX_+_15114892 | 0.83 |
ENSRNOT00000078168
|
Wdr13
|
WD repeat domain 13 |
| chr6_-_142534112 | 0.82 |
ENSRNOT00000073598
|
Ighv1-49
|
immunoglobulin heavy variable 1-49 |
| chr3_+_20993205 | 0.81 |
ENSRNOT00000010465
|
Olr419
|
olfactory receptor 419 |
| chr6_+_58388333 | 0.79 |
ENSRNOT00000036337
|
LOC680646
|
similar to 40S ribosomal protein S2 |
| chr3_-_139735397 | 0.75 |
ENSRNOT00000088315
|
AABR07054088.1
|
|
| chr7_+_140107488 | 0.66 |
ENSRNOT00000043467
|
Olr1111
|
olfactory receptor 1111 |
| chrX_+_20216587 | 0.65 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr20_+_5527181 | 0.60 |
ENSRNOT00000091364
|
Phf1
|
PHD finger protein 1 |
| chr3_-_78340788 | 0.54 |
ENSRNOT00000041870
|
Olr713
|
olfactory receptor 713 |
| chr3_-_103043741 | 0.47 |
ENSRNOT00000052294
|
Olr775
|
olfactory receptor 775 |
| chrX_-_81817101 | 0.47 |
ENSRNOT00000003730
|
AABR07039648.1
|
|
| chr15_+_32160364 | 0.43 |
ENSRNOT00000072888
|
AABR07017875.1
|
|
| chr1_+_68221395 | 0.36 |
ENSRNOT00000075596
|
Vom1r29
|
vomeronasal 1 receptor 29 |
| chr1_-_169892850 | 0.36 |
ENSRNOT00000037868
|
Olr183
|
olfactory receptor 183 |
| chr13_+_92412612 | 0.36 |
ENSRNOT00000004728
|
Olr1601
|
olfactory receptor 1601 |
| chr6_+_115170865 | 0.29 |
ENSRNOT00000005671
|
Tshr
|
thyroid stimulating hormone receptor |
| chr1_-_166912524 | 0.26 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr10_-_53595827 | 0.20 |
ENSRNOT00000004560
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr20_+_1861272 | 0.14 |
ENSRNOT00000044570
|
Olr1744
|
olfactory receptor 1744 |
| chr8_+_43733545 | 0.12 |
ENSRNOT00000082000
|
Olr1326
|
olfactory receptor 1326 |
| chr19_+_28377249 | 0.12 |
ENSRNOT00000059668
|
Vom1r21
|
vomeronasal 1 receptor 21 |
| chr3_-_166993940 | 0.09 |
ENSRNOT00000034669
|
Zfp217
|
zinc finger protein 217 |
| chr14_-_45165207 | 0.09 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr5_-_151029233 | 0.06 |
ENSRNOT00000089155
|
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
| chr20_+_1781320 | 0.04 |
ENSRNOT00000040388
|
Olr1737
|
olfactory receptor 1737 |
| chr3_-_166994286 | 0.02 |
ENSRNOT00000081593
|
Zfp217
|
zinc finger protein 217 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 41.9 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 6.5 | 32.7 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 5.9 | 41.3 | GO:0008228 | opsonization(GO:0008228) |
| 3.6 | 10.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 2.3 | 6.9 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 2.0 | 6.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.7 | 6.8 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.7 | 8.3 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.7 | 5.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.5 | 7.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 1.4 | 10.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 1.3 | 4.0 | GO:0001802 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.9 | 4.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.9 | 11.4 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.6 | 1.9 | GO:0035600 | tRNA methylthiolation(GO:0035600) |
| 0.6 | 5.0 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.5 | 1.5 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.5 | 1.9 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 1.3 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.4 | 3.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 3.3 | GO:0061450 | nail development(GO:0035878) trophoblast cell migration(GO:0061450) |
| 0.3 | 27.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.3 | 2.3 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.2 | 5.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 3.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 1.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 2.8 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 4.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.3 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.1 | 1.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 2.3 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 3.6 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.8 | 8.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 2.3 | 32.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 2.0 | 6.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 1.5 | 41.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.2 | 41.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.6 | 5.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.3 | 1.2 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.2 | 1.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 4.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 3.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 27.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 1.5 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 4.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 6.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 5.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 4.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 5.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.0 | GO:0005840 | ribosome(GO:0005840) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 2.3 | 6.9 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 1.7 | 41.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.5 | 6.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.4 | 8.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.3 | 10.0 | GO:0008430 | selenium binding(GO:0008430) |
| 1.1 | 4.4 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.0 | 5.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.8 | 4.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.7 | 3.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 1.9 | GO:0035596 | methylthiotransferase activity(GO:0035596) transferase activity, transferring alkylthio groups(GO:0050497) |
| 0.6 | 27.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.6 | 6.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 11.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.4 | 3.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.4 | 1.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.2 | 5.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 3.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 41.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 32.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.1 | 1.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 5.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 7.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 3.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 41.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.5 | 41.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 32.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 3.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 6.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 53.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 6.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 5.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 4.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 4.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 6.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 4.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 3.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |