Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
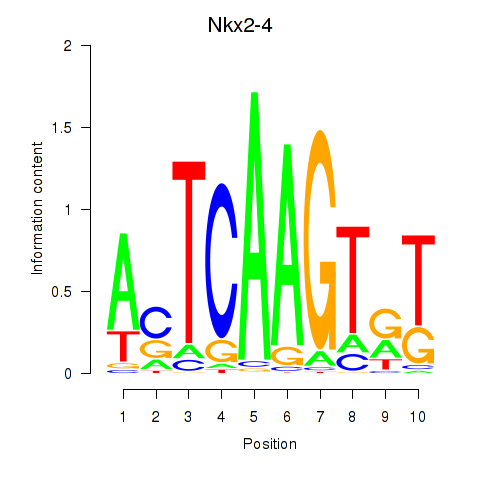
Results for Nkx2-4
Z-value: 0.51
Transcription factors associated with Nkx2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-4
|
ENSRNOG00000012647 | NK2 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-4 | rn6_v1_chr3_-_141305471_141305471 | -0.03 | 5.8e-01 | Click! |
Activity profile of Nkx2-4 motif
Sorted Z-values of Nkx2-4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_76789298 | 13.39 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr7_+_136182224 | 12.72 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr14_-_85191557 | 10.21 |
ENSRNOT00000011604
|
Nefh
|
neurofilament heavy |
| chr15_-_14737704 | 9.54 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr1_-_224698514 | 8.81 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr2_+_144861455 | 8.71 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr2_-_235177275 | 8.06 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr4_+_64088900 | 7.57 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr5_+_149077412 | 7.51 |
ENSRNOT00000014666
|
Matn1
|
matrilin 1, cartilage matrix protein |
| chr2_+_68820615 | 7.51 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr2_+_68821004 | 7.43 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr9_+_47281961 | 7.04 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr9_-_28732919 | 6.56 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr17_-_61332391 | 6.46 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr7_+_123510804 | 6.38 |
ENSRNOT00000010491
|
Sept3
|
septin 3 |
| chr6_+_43001948 | 6.27 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr14_+_113867209 | 6.10 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr4_-_49422824 | 5.93 |
ENSRNOT00000077889
|
Fam3c
|
family with sequence similarity 3, member C |
| chr18_+_29972808 | 5.54 |
ENSRNOT00000074051
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_+_100161872 | 5.35 |
ENSRNOT00000025777
|
Klk1c3
|
kallikrein 1-related peptidase C3 |
| chr18_+_29993361 | 4.75 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_-_100671074 | 4.73 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chr16_-_82099991 | 4.15 |
ENSRNOT00000089447
|
Atp11a
|
ATPase phospholipid transporting 11A |
| chr14_-_14390699 | 4.14 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr16_-_55132191 | 4.06 |
ENSRNOT00000017025
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
| chr9_+_82596355 | 3.80 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr5_+_78384444 | 3.44 |
ENSRNOT00000034978
|
LOC500475
|
similar to hypothetical protein 4933430I17 |
| chr1_+_101688297 | 3.33 |
ENSRNOT00000091606
|
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr8_-_55162421 | 3.17 |
ENSRNOT00000083102
|
Dixdc1
|
DIX domain containing 1 |
| chr3_-_9936352 | 3.11 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr2_+_72006099 | 2.92 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr10_-_66858598 | 2.90 |
ENSRNOT00000018898
|
Evi2b
|
ecotropic viral integration site 2B |
| chr4_-_160662974 | 2.86 |
ENSRNOT00000089199
|
Tspan9
|
tetraspanin 9 |
| chr2_+_95320283 | 2.71 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr19_-_10358695 | 2.69 |
ENSRNOT00000019770
|
Katnb1
|
katanin regulatory subunit B1 |
| chr3_-_141411170 | 2.65 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr8_+_107508351 | 2.61 |
ENSRNOT00000081594
|
Cep70
|
centrosomal protein 70 |
| chr15_-_60289763 | 2.61 |
ENSRNOT00000038579
|
Fam216b
|
family with sequence similarity 216, member B |
| chr4_-_22425381 | 2.45 |
ENSRNOT00000011166
ENSRNOT00000037712 |
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr4_-_129430251 | 2.41 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr18_-_63394690 | 2.33 |
ENSRNOT00000090078
ENSRNOT00000029431 |
Cep76
|
centrosomal protein 76 |
| chr3_+_19495628 | 2.28 |
ENSRNOT00000077990
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr3_+_2396143 | 2.19 |
ENSRNOT00000012388
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr3_-_93216495 | 2.08 |
ENSRNOT00000010580
|
Ehf
|
ets homologous factor |
| chr17_-_29895386 | 2.05 |
ENSRNOT00000048757
|
Cdyl
|
chromodomain Y-like |
| chr1_-_67134827 | 2.01 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr19_+_57047830 | 1.90 |
ENSRNOT00000080860
|
Galnt2
|
polypeptide N-acetylgalactosaminyltransferase 2 |
| chr4_-_155421998 | 1.78 |
ENSRNOT00000041552
|
Gdf3
|
growth differentiation factor 3 |
| chr2_+_22910236 | 1.77 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr15_-_14510828 | 1.71 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr3_+_19690016 | 1.69 |
ENSRNOT00000085460
|
AABR07051707.1
|
|
| chr14_+_72889038 | 1.59 |
ENSRNOT00000084261
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr16_-_8564379 | 1.58 |
ENSRNOT00000060975
|
LOC680885
|
hypothetical protein LOC680885 |
| chr1_-_242765807 | 1.42 |
ENSRNOT00000020763
|
Pgm5
|
phosphoglucomutase 5 |
| chr10_-_19574094 | 1.40 |
ENSRNOT00000059810
|
Dock2
|
dedicator of cytokinesis 2 |
| chr5_-_139853247 | 1.35 |
ENSRNOT00000056644
|
Exo5
|
exonuclease 5 |
| chr1_-_82004538 | 1.31 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr14_-_74095146 | 1.30 |
ENSRNOT00000049313
|
LOC679766
|
similar to developmental pluripotency-associated 3 |
| chr4_+_120672152 | 1.28 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr12_-_13133219 | 1.26 |
ENSRNOT00000092292
|
Daglb
|
diacylglycerol lipase, beta |
| chr7_-_9104719 | 1.16 |
ENSRNOT00000088247
|
LOC103692758
|
olfactory receptor 6C74-like |
| chr4_+_82783686 | 1.15 |
ENSRNOT00000041988
|
Tax1bp1
|
Tax1 binding protein 1 |
| chr20_-_5227620 | 1.14 |
ENSRNOT00000086240
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr4_-_68819872 | 1.03 |
ENSRNOT00000033265
ENSRNOT00000081884 |
Clec5a
|
C-type lectin domain family 5, member A |
| chr12_-_19114399 | 0.96 |
ENSRNOT00000073099
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr3_-_173944396 | 0.95 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr14_-_107160334 | 0.86 |
ENSRNOT00000012131
|
Ehbp1
|
EH domain binding protein 1 |
| chr10_-_70646495 | 0.80 |
ENSRNOT00000071218
|
Gas2l2
|
growth arrest-specific 2 like 2 |
| chr1_-_155955173 | 0.79 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr9_-_17163170 | 0.73 |
ENSRNOT00000025921
|
Xpo5
|
exportin 5 |
| chr6_+_10306405 | 0.67 |
ENSRNOT00000090420
ENSRNOT00000080507 |
Epas1
|
endothelial PAS domain protein 1 |
| chr8_+_1502877 | 0.67 |
ENSRNOT00000034482
|
Msantd4
|
Myb/SANT DNA binding domain containing 4 with coiled-coils |
| chr10_+_90984227 | 0.65 |
ENSRNOT00000003890
ENSRNOT00000093707 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr8_+_96551245 | 0.63 |
ENSRNOT00000039850
|
Bcl2a1
|
BCL2-related protein A1 |
| chr2_-_240866689 | 0.58 |
ENSRNOT00000036838
|
Nfkb1
|
nuclear factor kappa B subunit 1 |
| chr7_+_107467260 | 0.50 |
ENSRNOT00000009240
ENSRNOT00000077382 |
Tg
|
thyroglobulin |
| chr2_-_213178632 | 0.44 |
ENSRNOT00000023079
|
LOC108348173
|
protein arginine N-methyltransferase 6 |
| chr1_+_60248695 | 0.31 |
ENSRNOT00000079558
|
Vom1r9
|
vomeronasal 1 receptor 9 |
| chr8_-_20173107 | 0.30 |
ENSRNOT00000045215
|
Olr1163
|
olfactory receptor 1163 |
| chr13_-_26769374 | 0.27 |
ENSRNOT00000003768
|
Bcl2
|
BCL2, apoptosis regulator |
| chr1_+_170147300 | 0.26 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr3_-_74567685 | 0.25 |
ENSRNOT00000068048
|
Olr532
|
olfactory receptor 532 |
| chr1_+_75233703 | 0.24 |
ENSRNOT00000064178
|
LOC100911727
|
DNA ligase 1-like |
| chr7_-_130170609 | 0.20 |
ENSRNOT00000088030
|
Dennd6b
|
DENN domain containing 6B |
| chr20_+_1736377 | 0.17 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr3_+_18805220 | 0.17 |
ENSRNOT00000071216
|
Igkv4-86
|
immunoglobulin kappa variable 4-86 |
| chr6_+_136084605 | 0.14 |
ENSRNOT00000092806
|
Mark3
|
microtubule affinity regulating kinase 3 |
| chr5_+_118418799 | 0.12 |
ENSRNOT00000012052
|
Alg6
|
ALG6, alpha-1,3-glucosyltransferase |
| chr1_-_44615760 | 0.09 |
ENSRNOT00000022148
|
Nox3
|
NADPH oxidase 3 |
| chr7_+_140742418 | 0.06 |
ENSRNOT00000089211
|
Prph
|
peripherin |
| chr1_-_48686713 | 0.03 |
ENSRNOT00000091196
|
AABR07001516.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.2 | 6.6 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 2.0 | 6.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.9 | 14.9 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 1.5 | 7.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.1 | 7.5 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.9 | 2.7 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.9 | 2.7 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.9 | 2.7 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.8 | 8.8 | GO:0015747 | urate transport(GO:0015747) |
| 0.6 | 2.5 | GO:1905235 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.5 | 7.0 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 1.6 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.5 | 1.9 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.5 | 3.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.4 | 1.3 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.3 | 1.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.3 | 4.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 4.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.2 | 0.7 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 1.0 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.2 | 8.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 0.6 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.2 | 1.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.2 | 1.4 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) establishment of T cell polarity(GO:0001768) |
| 0.2 | 4.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 1.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.2 | 4.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 2.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.7 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 1.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 | 1.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.3 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 0.7 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.0 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 7.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.4 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.2 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 1.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 2.1 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 3.3 | GO:0007623 | circadian rhythm(GO:0007623) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.6 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.3 | 6.4 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 1.3 | 7.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.2 | 4.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.9 | 10.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.5 | 1.6 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.4 | 9.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 2.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 4.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 2.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.7 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 1.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 8.4 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.7 | GO:0044450 | microtubule organizing center part(GO:0044450) |
| 0.0 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 14.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 7.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 5.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 10.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.9 | 2.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.9 | 8.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.9 | 14.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.8 | 4.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.7 | 7.0 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 10.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 2.5 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.4 | 1.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 4.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 6.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.3 | 1.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 4.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 2.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.6 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 1.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 3.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 7.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 6.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 5.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 14.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 6.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 6.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 5.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 14.9 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.5 | 6.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 7.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 4.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 7.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |