Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
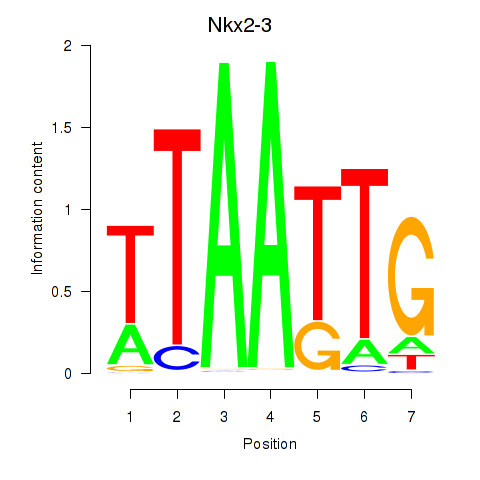
Results for Nkx2-3
Z-value: 1.10
Transcription factors associated with Nkx2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-3
|
ENSRNOG00000016656 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-3 | rn6_v1_chr1_+_263367266_263367266 | -0.32 | 6.6e-09 | Click! |
Activity profile of Nkx2-3 motif
Sorted Z-values of Nkx2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_118197217 | 72.62 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr14_+_39663421 | 60.70 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr6_+_58468155 | 43.33 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr9_-_30844199 | 42.25 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr9_+_73418607 | 40.00 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr9_+_73334618 | 37.42 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr1_-_70235091 | 35.84 |
ENSRNOT00000075745
|
Apeg3
|
antisense paternally expressed gene 3 |
| chr14_+_99529284 | 35.03 |
ENSRNOT00000006897
ENSRNOT00000067134 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr8_-_82533689 | 32.86 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr6_+_64789940 | 32.74 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr3_+_48082935 | 32.65 |
ENSRNOT00000087711
ENSRNOT00000067545 |
Slc4a10
|
solute carrier family 4 member 10 |
| chrX_+_107496072 | 32.63 |
ENSRNOT00000003283
|
Plp1
|
proteolipid protein 1 |
| chr4_-_55011415 | 30.52 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr2_+_144861455 | 30.26 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chrX_+_114930457 | 30.12 |
ENSRNOT00000089141
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr2_-_35104963 | 29.29 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr3_-_51612397 | 29.14 |
ENSRNOT00000081401
|
Scn3a
|
sodium voltage-gated channel alpha subunit 3 |
| chr2_+_136993208 | 28.09 |
ENSRNOT00000040187
ENSRNOT00000066542 |
Pcdh10
|
protocadherin 10 |
| chr10_-_108691367 | 28.02 |
ENSRNOT00000005067
|
Nptx1
|
neuronal pentraxin 1 |
| chr4_+_172942020 | 28.01 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr9_-_63641400 | 27.78 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr15_+_56666012 | 27.02 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr1_+_256955652 | 26.17 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr3_+_177310753 | 25.59 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr17_-_9762813 | 25.43 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr3_-_26056818 | 25.31 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr18_-_24182012 | 24.57 |
ENSRNOT00000023594
|
Syt4
|
synaptotagmin 4 |
| chr5_-_130085838 | 24.38 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr4_-_184096806 | 23.42 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr8_+_82038967 | 23.39 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr15_+_87886783 | 22.16 |
ENSRNOT00000065710
|
Slain1
|
SLAIN motif family, member 1 |
| chr4_-_15859132 | 21.66 |
ENSRNOT00000082161
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr6_+_73553210 | 21.05 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr2_-_5577369 | 20.86 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr10_-_82229140 | 20.60 |
ENSRNOT00000004400
|
Epn3
|
epsin 3 |
| chr3_-_105470475 | 20.59 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr10_-_18942540 | 20.58 |
ENSRNOT00000007187
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr5_+_119727839 | 19.04 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr15_+_59678165 | 18.77 |
ENSRNOT00000074868
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr1_-_93949187 | 18.70 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr13_+_87986240 | 18.58 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr14_-_46153212 | 18.54 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr20_+_25990656 | 18.48 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr5_-_65073012 | 18.33 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr18_+_27632786 | 18.29 |
ENSRNOT00000073564
ENSRNOT00000078969 |
Reep2
|
receptor accessory protein 2 |
| chr10_-_66848388 | 17.91 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr17_-_63994169 | 17.72 |
ENSRNOT00000075651
|
Chrm3
|
cholinergic receptor, muscarinic 3 |
| chrX_+_6273733 | 17.43 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr4_+_110699557 | 17.20 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr18_+_54108493 | 17.08 |
ENSRNOT00000029239
|
RGD1312005
|
similar to DD1 |
| chrX_-_82743753 | 17.05 |
ENSRNOT00000003512
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr9_+_62291405 | 16.74 |
ENSRNOT00000051463
|
Plcl1
|
phospholipase C-like 1 |
| chr9_+_86874685 | 16.60 |
ENSRNOT00000041337
|
NEWGENE_1305560
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr7_+_44009069 | 16.49 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr16_-_73152921 | 16.49 |
ENSRNOT00000048602
|
Zmat4
|
zinc finger, matrin type 4 |
| chr7_-_52404774 | 16.32 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr2_-_96509424 | 16.14 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr4_+_64088900 | 16.12 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr9_+_47281961 | 15.83 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr1_-_173764246 | 15.42 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr7_+_42304534 | 15.42 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr10_-_91986632 | 15.28 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr2_-_188559882 | 15.19 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr5_-_93244202 | 14.91 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr9_+_60039297 | 14.88 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr4_+_96562725 | 14.78 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr3_-_45169118 | 14.40 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr1_+_61786900 | 14.36 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr9_+_25410669 | 13.70 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr18_+_29999290 | 13.50 |
ENSRNOT00000027372
|
Pcdha4
|
protocadherin alpha 4 |
| chr6_+_106496992 | 13.42 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr18_+_30424814 | 13.39 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr16_+_22361998 | 13.36 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr5_-_39215102 | 13.10 |
ENSRNOT00000050653
|
Klhl32
|
kelch-like family member 32 |
| chr2_+_72006099 | 12.87 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr10_-_86004096 | 12.81 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr14_+_37130743 | 12.77 |
ENSRNOT00000031756
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr5_+_25349928 | 12.54 |
ENSRNOT00000020826
|
Gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr8_-_21384131 | 12.19 |
ENSRNOT00000071010
|
Zfp560
|
zinc finger protein 560 |
| chr1_-_43638161 | 12.17 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr12_-_44520341 | 12.04 |
ENSRNOT00000066810
|
Nos1
|
nitric oxide synthase 1 |
| chr1_-_90520344 | 11.95 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr3_+_128828331 | 11.60 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr6_-_114476723 | 11.30 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr10_-_74679858 | 11.14 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr13_-_76049363 | 11.12 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chrX_-_121731543 | 11.06 |
ENSRNOT00000018788
|
Klhl13
|
kelch-like family member 13 |
| chr9_-_121725716 | 10.81 |
ENSRNOT00000087405
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr3_+_38559914 | 10.80 |
ENSRNOT00000085545
|
Fmnl2
|
formin-like 2 |
| chr18_-_26656879 | 10.77 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_+_11658568 | 10.53 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr11_-_768644 | 10.51 |
ENSRNOT00000090197
|
Epha3
|
Eph receptor A3 |
| chr18_+_30820321 | 10.47 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr14_+_104250617 | 10.27 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr6_+_76745981 | 10.23 |
ENSRNOT00000076784
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr16_-_6245644 | 10.14 |
ENSRNOT00000040759
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_+_4252496 | 9.98 |
ENSRNOT00000071535
|
RGD1560883
|
similar to KIAA0825 protein |
| chr13_+_106463368 | 9.92 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chrX_+_25016401 | 9.73 |
ENSRNOT00000059270
|
Clcn4
|
chloride voltage-gated channel 4 |
| chr8_+_122076759 | 9.70 |
ENSRNOT00000012545
|
Clasp2
|
cytoplasmic linker associated protein 2 |
| chr6_-_67084234 | 9.43 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr9_+_51302151 | 9.22 |
ENSRNOT00000085908
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_153803349 | 9.11 |
ENSRNOT00000088565
|
Mme
|
membrane metallo-endopeptidase |
| chr1_+_89215266 | 9.04 |
ENSRNOT00000093612
ENSRNOT00000084799 |
Dmkn
|
dermokine |
| chr2_-_199971965 | 8.95 |
ENSRNOT00000087026
|
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr16_-_2602103 | 8.83 |
ENSRNOT00000018413
|
Appl1
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 |
| chr17_-_81187739 | 8.70 |
ENSRNOT00000063911
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chrX_-_10031167 | 8.62 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chrX_-_64908682 | 8.58 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr3_-_141411170 | 8.48 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr5_+_146656049 | 8.38 |
ENSRNOT00000036029
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr12_+_2140203 | 8.34 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr14_-_79988110 | 8.34 |
ENSRNOT00000090597
|
Afap1
|
actin filament associated protein 1 |
| chr19_+_46733633 | 8.21 |
ENSRNOT00000016307
|
Clec3a
|
C-type lectin domain family 3, member A |
| chrX_+_63542191 | 8.21 |
ENSRNOT00000073955
|
Apoo
|
apolipoprotein O |
| chrX_+_40460047 | 8.18 |
ENSRNOT00000010970
|
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr7_-_40316532 | 8.14 |
ENSRNOT00000083347
|
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr8_-_12993651 | 8.00 |
ENSRNOT00000033932
|
Kdm4d
|
lysine demethylase 4D |
| chr2_+_204427608 | 7.79 |
ENSRNOT00000083374
|
Nhlh2
|
nescient helix loop helix 2 |
| chr13_+_98231326 | 7.72 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_+_145174876 | 7.66 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr13_-_102857551 | 7.63 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chrX_-_153878806 | 7.48 |
ENSRNOT00000087990
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr1_+_53220397 | 7.44 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr4_+_2055615 | 7.34 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr11_-_782954 | 7.33 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr2_-_184263564 | 7.28 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr5_-_106923132 | 7.25 |
ENSRNOT00000046207
|
Ifna4
|
interferon, alpha 4 |
| chr5_-_99033107 | 7.22 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr3_-_21904133 | 7.15 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr17_+_11683862 | 7.13 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr19_-_56983807 | 7.07 |
ENSRNOT00000043529
|
Cox6c-ps1
|
cytochrome c oxidase subunit VIc, pseudogene |
| chrX_-_25590048 | 6.95 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr10_-_45812641 | 6.89 |
ENSRNOT00000038951
|
Zfp867
|
zinc finger protein 867 |
| chr19_+_39229754 | 6.84 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chrX_+_158835811 | 6.64 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr2_+_266315036 | 6.50 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr3_-_77227036 | 6.41 |
ENSRNOT00000072638
|
LOC103691843
|
olfactory receptor 4P4-like |
| chr5_+_58995249 | 6.39 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr6_+_22696397 | 6.20 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chr2_-_199354793 | 6.18 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr4_+_87608301 | 6.16 |
ENSRNOT00000058702
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr6_-_51018050 | 6.15 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr20_-_27117663 | 5.97 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chrX_+_9956370 | 5.96 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr14_-_79987915 | 5.95 |
ENSRNOT00000089006
|
Afap1
|
actin filament associated protein 1 |
| chr18_-_75207306 | 5.91 |
ENSRNOT00000021717
|
Setbp1
|
SET binding protein 1 |
| chr19_-_49510901 | 5.84 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chrX_-_40086870 | 5.76 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr8_-_18037774 | 5.71 |
ENSRNOT00000074948
|
Olr1137
|
olfactory receptor 1137 |
| chr3_-_64364101 | 5.63 |
ENSRNOT00000089065
|
Zfp385b
|
zinc finger protein 385B |
| chr15_-_37831031 | 5.59 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr10_+_74436534 | 5.48 |
ENSRNOT00000008298
|
Trim37
|
tripartite motif-containing 37 |
| chr19_+_39357791 | 5.41 |
ENSRNOT00000086435
ENSRNOT00000015006 |
Cyb5b
|
cytochrome b5 type B |
| chr1_+_217459215 | 5.39 |
ENSRNOT00000092386
ENSRNOT00000092357 |
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_+_52127399 | 5.38 |
ENSRNOT00000052392
|
Cadm1
|
cell adhesion molecule 1 |
| chr2_+_86891092 | 5.36 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr7_-_55604403 | 5.34 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr7_-_4216833 | 5.31 |
ENSRNOT00000003174
|
Olr982
|
olfactory receptor 982 |
| chr2_+_122865100 | 5.28 |
ENSRNOT00000067090
|
Rslcan18
|
regulator of sex-limitation candidate 18 |
| chr4_-_167068838 | 5.13 |
ENSRNOT00000029713
|
Tas2r125
|
taste receptor, type 2, member 125 |
| chr7_+_42269784 | 5.13 |
ENSRNOT00000008471
ENSRNOT00000007231 |
Kitlg
|
KIT ligand |
| chr14_+_62309299 | 5.12 |
ENSRNOT00000048309
|
Vom1r26
|
vomeronasal 1 receptor 26 |
| chrX_+_136488691 | 5.08 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr9_+_66888393 | 5.05 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr2_+_186980992 | 5.03 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr1_-_99632845 | 4.99 |
ENSRNOT00000049123
|
LOC103691107
|
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 pseudogene |
| chr6_-_106971250 | 4.98 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr2_+_177651241 | 4.97 |
ENSRNOT00000047593
|
AABR07011951.1
|
|
| chr1_-_204275803 | 4.83 |
ENSRNOT00000066711
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chrX_+_54062935 | 4.81 |
ENSRNOT00000004854
|
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr1_-_149633029 | 4.81 |
ENSRNOT00000048861
|
LOC100910049
|
olfactory receptor 14A2-like |
| chr2_-_45518502 | 4.79 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr7_+_18120715 | 4.78 |
ENSRNOT00000068151
|
Vom1r108
|
vomeronasal 1 receptor 108 |
| chr8_+_52127632 | 4.76 |
ENSRNOT00000079797
|
Cadm1
|
cell adhesion molecule 1 |
| chr1_-_219369587 | 4.67 |
ENSRNOT00000000599
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr3_+_168124673 | 4.62 |
ENSRNOT00000070809
|
Pfdn4
|
prefoldin subunit 4 |
| chr15_-_95514259 | 4.60 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr1_+_95397991 | 4.58 |
ENSRNOT00000039649
|
Zfp939
|
zinc finger protein 939 |
| chr12_+_38377034 | 4.55 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr19_+_25043680 | 4.52 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr14_+_104480662 | 4.51 |
ENSRNOT00000078710
|
Rab1a
|
RAB1A, member RAS oncogene family |
| chr3_-_102672294 | 4.48 |
ENSRNOT00000030909
|
Olr760
|
olfactory receptor 760 |
| chr10_+_94260197 | 4.37 |
ENSRNOT00000063973
|
Taco1
|
translational activator of cytochrome c oxidase I |
| chr4_+_31333970 | 4.34 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr6_+_28382962 | 4.31 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr2_-_173836854 | 4.21 |
ENSRNOT00000074278
|
Wdr49
|
WD repeat domain 49 |
| chr9_-_66019065 | 4.18 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr8_+_100260049 | 4.08 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr6_+_72124417 | 3.96 |
ENSRNOT00000040548
|
Scfd1
|
sec1 family domain containing 1 |
| chr2_-_177950517 | 3.77 |
ENSRNOT00000012792
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr4_+_21872856 | 3.73 |
ENSRNOT00000044810
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr2_+_256609787 | 3.71 |
ENSRNOT00000047564
ENSRNOT00000088437 |
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr4_+_88694583 | 3.58 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chrX_-_42329232 | 3.41 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chrX_+_69730242 | 3.35 |
ENSRNOT00000075980
ENSRNOT00000076425 |
Eda
|
ectodysplasin-A |
| chr2_+_243018975 | 3.32 |
ENSRNOT00000081989
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14 |
| chr5_-_147584038 | 3.23 |
ENSRNOT00000010983
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr7_-_95310005 | 3.21 |
ENSRNOT00000005815
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr6_-_105261549 | 3.20 |
ENSRNOT00000009122
|
Synj2bp
|
synaptojanin 2 binding protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.2 | 99.6 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 8.5 | 25.4 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 7.0 | 35.0 | GO:0070350 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 6.8 | 33.8 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 5.9 | 17.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 5.8 | 23.4 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 5.3 | 21.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 5.1 | 25.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 4.9 | 24.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 4.3 | 17.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 4.1 | 20.5 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 3.9 | 15.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 3.8 | 15.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 3.8 | 30.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 3.6 | 32.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 3.6 | 43.3 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 3.6 | 21.7 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 3.5 | 28.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 3.5 | 10.5 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 3.0 | 32.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 2.9 | 29.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 2.9 | 17.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 2.9 | 8.7 | GO:0071529 | cementum mineralization(GO:0071529) |
| 2.9 | 22.8 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 2.8 | 8.5 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 2.8 | 27.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 2.8 | 30.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 2.7 | 8.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 2.7 | 10.8 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 2.7 | 8.0 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 2.6 | 10.5 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 2.5 | 28.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 2.4 | 7.3 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 2.4 | 7.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 2.4 | 94.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 2.3 | 25.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 2.3 | 13.7 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 2.3 | 9.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 2.3 | 9.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 2.2 | 6.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 2.1 | 20.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 2.1 | 18.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 2.1 | 6.2 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 2.1 | 6.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 2.0 | 10.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 2.0 | 6.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 1.9 | 14.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.8 | 14.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.8 | 77.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 1.8 | 8.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.7 | 31.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 1.7 | 8.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.5 | 18.5 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 1.5 | 15.2 | GO:0099612 | protein localization to axon(GO:0099612) |
| 1.5 | 3.0 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 1.5 | 14.9 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 1.4 | 5.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 1.3 | 15.8 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.3 | 17.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 1.3 | 18.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 1.3 | 30.1 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 1.3 | 11.3 | GO:0042447 | phenol-containing compound catabolic process(GO:0019336) hormone catabolic process(GO:0042447) |
| 1.2 | 20.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 1.2 | 4.7 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 1.1 | 2.3 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.1 | 2.3 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.1 | 3.4 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.0 | 3.1 | GO:0043132 | NAD transport(GO:0043132) |
| 1.0 | 6.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.9 | 4.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.9 | 10.0 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.9 | 30.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.8 | 4.8 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.8 | 7.7 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.8 | 3.8 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.7 | 11.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.7 | 2.0 | GO:0015692 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 0.6 | 7.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.6 | 17.9 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.6 | 7.2 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.6 | 5.4 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.6 | 4.2 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.6 | 10.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.6 | 4.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.5 | 2.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.5 | 1.6 | GO:0071611 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) negative regulation of interferon-alpha production(GO:0032687) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) |
| 0.5 | 8.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.5 | 2.3 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.5 | 5.0 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.4 | 18.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.4 | 4.0 | GO:1902902 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) negative regulation of autophagosome assembly(GO:1902902) |
| 0.4 | 2.7 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.4 | 9.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.4 | 3.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.4 | 24.4 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.4 | 1.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.3 | 2.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.3 | 10.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 1.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.3 | 2.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.3 | 4.5 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 1.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.3 | 37.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 7.5 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 | 3.2 | GO:0048312 | positive regulation of activin receptor signaling pathway(GO:0032927) intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 1.7 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.2 | 2.5 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 9.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.2 | 2.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 9.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.2 | 21.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.2 | 2.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 35.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.2 | 26.2 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.2 | 8.8 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.2 | 1.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 0.8 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.2 | 3.9 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 0.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 2.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.9 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.1 | 14.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 10.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.3 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.1 | 2.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 3.4 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 | 9.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 2.9 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.1 | 0.1 | GO:0038001 | paracrine signaling(GO:0038001) |
| 0.1 | 7.6 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.1 | 0.3 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 5.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 4.8 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.1 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 4.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 6.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 6.3 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.0 | 6.6 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 11.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 5.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 1.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.5 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.3 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 2.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 110.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 7.8 | 23.4 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 7.0 | 21.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 3.4 | 10.2 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 3.0 | 15.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 2.7 | 16.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 2.7 | 37.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 2.6 | 60.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 2.4 | 24.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 2.3 | 27.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.8 | 32.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.6 | 24.6 | GO:0031045 | dense core granule(GO:0031045) |
| 1.5 | 29.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.5 | 7.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.4 | 9.7 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 1.2 | 8.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.2 | 17.7 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 1.2 | 8.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.2 | 5.8 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.1 | 18.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.0 | 6.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.9 | 4.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 15.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.7 | 5.4 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.7 | 12.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.7 | 2.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.6 | 4.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.6 | 17.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 2.8 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 29.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.5 | 6.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.4 | 8.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 19.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.4 | 35.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 5.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 20.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.3 | 25.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 11.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 5.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 12.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 22.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 8.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 2.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 6.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 20.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 40.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 8.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 2.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 14.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 27.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 3.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 10.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 9.7 | GO:0031902 | early endosome membrane(GO:0031901) late endosome membrane(GO:0031902) |
| 0.1 | 120.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 3.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.9 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 6.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 41.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 9.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 6.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 12.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 10.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 33.1 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 1.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 6.6 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 1.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 8.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 7.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.2 | 99.6 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 12.1 | 60.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 7.8 | 23.4 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 7.6 | 30.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 7.2 | 21.7 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 6.8 | 33.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 5.1 | 25.4 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 4.9 | 39.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 4.9 | 24.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.5 | 13.4 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 4.4 | 30.8 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 3.8 | 87.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 3.6 | 32.7 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 3.2 | 3.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 2.9 | 8.7 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 2.7 | 32.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.6 | 10.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 2.5 | 9.9 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 2.5 | 29.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 2.4 | 12.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 2.3 | 21.0 | GO:0043495 | protein anchor(GO:0043495) |
| 2.1 | 38.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 1.8 | 18.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.8 | 17.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.7 | 30.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.6 | 4.8 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.6 | 15.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 1.5 | 7.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.3 | 28.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 1.2 | 4.7 | GO:0036004 | GAF domain binding(GO:0036004) |
| 1.1 | 29.1 | GO:0031402 | sodium ion binding(GO:0031402) |
| 1.1 | 4.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.1 | 10.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 1.0 | 6.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.0 | 4.1 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 1.0 | 3.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.9 | 24.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.9 | 7.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.9 | 24.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.8 | 2.4 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.8 | 8.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.7 | 7.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.7 | 2.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.7 | 15.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.6 | 3.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.6 | 3.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.6 | 7.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 14.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.6 | 13.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.6 | 1.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.5 | 8.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.5 | 9.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 3.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 10.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.4 | 6.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 17.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.4 | 5.4 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.3 | 2.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 2.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 1.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.3 | 10.2 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.3 | 0.9 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.3 | 6.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 34.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.3 | 0.9 | GO:0034512 | box C/D snoRNA binding(GO:0034512) |
| 0.3 | 2.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 0.8 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.3 | 5.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 10.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 7.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 4.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 8.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 14.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.2 | 2.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 2.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.9 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.2 | 4.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 5.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 5.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 0.7 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 5.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 48.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.2 | 24.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.2 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 1.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 2.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 9.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 17.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 59.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 10.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 1.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 2.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 6.2 | GO:0019199 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) transmembrane receptor protein kinase activity(GO:0019199) |
| 0.1 | 0.9 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 5.6 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 3.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 6.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 6.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 10.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 18.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 16.0 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.1 | 8.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 20.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 4.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 11.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 7.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 10.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 2.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 42.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 30.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 5.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 4.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 4.3 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 43.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 1.4 | 77.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.8 | 33.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.7 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.6 | 17.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.6 | 12.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 21.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.4 | 10.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.4 | 11.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.3 | 17.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 4.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 7.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 17.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 11.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 6.3 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 4.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 36.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 5.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 7.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 6.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 8.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 9.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 3.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 6.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 99.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 3.6 | 60.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.8 | 17.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 1.4 | 30.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.0 | 16.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.9 | 32.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.9 | 20.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.9 | 23.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.9 | 14.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.9 | 15.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.8 | 15.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.8 | 17.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.6 | 16.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.6 | 8.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.6 | 13.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 10.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.5 | 14.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 13.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 4.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.4 | 30.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.4 | 4.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 12.0 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.4 | 8.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.4 | 5.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 6.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 33.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 16.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 24.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 10.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 4.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 7.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 4.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 3.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.7 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 15.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 2.7 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 2.3 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 2.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 6.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |