Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
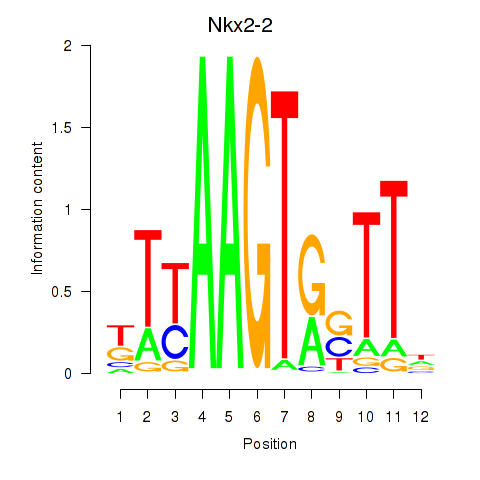
Results for Nkx2-2
Z-value: 0.90
Transcription factors associated with Nkx2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-2
|
ENSRNOG00000012728 | NK2 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-2 | rn6_v1_chr3_-_141411170_141411170 | 0.26 | 1.6e-06 | Click! |
Activity profile of Nkx2-2 motif
Sorted Z-values of Nkx2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_28654733 | 33.32 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chrX_+_78372257 | 29.42 |
ENSRNOT00000046164
|
Gpr174
|
G protein-coupled receptor 174 |
| chr1_-_104202591 | 27.71 |
ENSRNOT00000035512
|
E2f8
|
E2F transcription factor 8 |
| chr4_+_101882994 | 26.50 |
ENSRNOT00000087773
|
AABR07060963.1
|
|
| chr8_+_55603968 | 25.77 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr1_+_192379543 | 24.16 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr14_-_43992587 | 23.56 |
ENSRNOT00000003425
|
Rhoh
|
ras homolog family member H |
| chr13_+_91954138 | 20.00 |
ENSRNOT00000074608
ENSRNOT00000037655 |
Aim2
|
absent in melanoma 2 |
| chr17_+_44763598 | 19.71 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr3_+_16413080 | 19.11 |
ENSRNOT00000040386
|
LOC100912707
|
Ig kappa chain V19-17-like |
| chr1_-_227441442 | 18.78 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr9_-_92775816 | 17.67 |
ENSRNOT00000029635
|
RGD1563917
|
similar to Nuclear autoantigen Sp-100 (Speckled 100 kDa) |
| chr6_-_137664133 | 17.40 |
ENSRNOT00000018613
|
Gpr132
|
G protein-coupled receptor 132 |
| chr1_+_227670159 | 15.74 |
ENSRNOT00000072077
|
Ms4a6c
|
membrane-spanning 4-domains, subfamily A, member 6C |
| chr13_-_74520634 | 15.42 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr4_+_145489869 | 14.65 |
ENSRNOT00000082618
|
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr17_-_85557939 | 14.40 |
ENSRNOT00000022375
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase type 2 alpha |
| chr11_+_85263536 | 14.04 |
ENSRNOT00000046465
|
AABR07034729.1
|
|
| chr7_-_104556419 | 13.94 |
ENSRNOT00000090784
|
Fam49b
|
family with sequence similarity 49, member B |
| chr4_-_28310178 | 13.23 |
ENSRNOT00000084021
|
RGD1563091
|
similar to OEF2 |
| chr14_+_44889287 | 12.69 |
ENSRNOT00000091312
ENSRNOT00000032273 |
Tmem156
|
transmembrane protein 156 |
| chr8_-_132919110 | 12.35 |
ENSRNOT00000008747
|
Xcr1
|
X-C motif chemokine receptor 1 |
| chr4_+_101639641 | 12.15 |
ENSRNOT00000058282
|
AABR07060952.1
|
|
| chr19_+_56220755 | 12.13 |
ENSRNOT00000023452
|
Tubb3
|
tubulin, beta 3 class III |
| chrX_+_78259409 | 11.90 |
ENSRNOT00000049779
|
RGD1560455
|
similar to RIKEN cDNA A630033H20 gene |
| chr1_+_98398660 | 11.62 |
ENSRNOT00000047473
|
Cd33
|
CD33 molecule |
| chr8_+_79054237 | 11.59 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr7_-_54778848 | 11.41 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr12_-_2826378 | 11.37 |
ENSRNOT00000061749
|
Clec4m
|
C-type lectin domain family 4 member M |
| chr14_+_88549947 | 10.71 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr2_-_149444548 | 10.60 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr9_-_101038625 | 10.54 |
ENSRNOT00000025743
|
Pdcd1
|
programmed cell death 1 |
| chr11_+_20474483 | 10.53 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr14_+_100135271 | 9.89 |
ENSRNOT00000032965
|
Fbxo48
|
F-box protein 48 |
| chr13_-_95348913 | 9.87 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr10_-_90994437 | 9.80 |
ENSRNOT00000093167
|
Gfap
|
glial fibrillary acidic protein |
| chr7_+_133856101 | 9.53 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr20_-_14620019 | 9.49 |
ENSRNOT00000001779
|
Gnaz
|
G protein subunit alpha z |
| chr15_+_31642169 | 9.45 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chrX_-_138435391 | 9.43 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr9_-_27761733 | 9.30 |
ENSRNOT00000040034
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr7_+_126736732 | 9.19 |
ENSRNOT00000022012
|
Gtse1
|
G-2 and S-phase expressed 1 |
| chr9_-_27761365 | 9.09 |
ENSRNOT00000018552
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr2_+_34312766 | 8.98 |
ENSRNOT00000060962
|
Cenpk
|
centromere protein K |
| chr6_+_8669722 | 8.87 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr1_-_198476476 | 8.68 |
ENSRNOT00000027525
|
Kif22
|
kinesin family member 22 |
| chr4_-_135069970 | 8.66 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr13_-_91981432 | 8.57 |
ENSRNOT00000004637
|
AABR07021804.1
|
|
| chr4_+_146276862 | 8.37 |
ENSRNOT00000009705
|
Slc6a1
|
solute carrier family 6 member 1 |
| chr9_-_61418679 | 8.36 |
ENSRNOT00000078800
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr15_+_4850122 | 7.89 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr17_-_42422053 | 7.77 |
ENSRNOT00000048298
|
Fam65b
|
family with sequence similarity 65, member B |
| chr3_+_19174027 | 7.75 |
ENSRNOT00000074445
|
AABR07051678.1
|
|
| chr1_+_173252058 | 7.58 |
ENSRNOT00000073421
|
LOC499229
|
similar to very large inducible GTPase 1 isoform A |
| chr7_-_142063212 | 7.53 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr4_-_180234804 | 7.43 |
ENSRNOT00000070957
|
Bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr19_-_9843673 | 7.17 |
ENSRNOT00000015795
|
Gins3
|
GINS complex subunit 3 |
| chr20_-_22459025 | 7.07 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr5_+_122019301 | 7.07 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr4_-_164304532 | 6.87 |
ENSRNOT00000089484
|
Ly49i5
|
Ly49 inhibitory receptor 5 |
| chr5_+_173542358 | 6.82 |
ENSRNOT00000027347
|
RGD1311517
|
similar to RIKEN cDNA 9430015G10 |
| chr5_+_169288871 | 6.69 |
ENSRNOT00000055466
|
Tnfrsf25
|
TNF receptor superfamily member 25 |
| chr1_+_101249522 | 6.68 |
ENSRNOT00000033882
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr9_-_101319845 | 6.61 |
ENSRNOT00000074915
|
NEWGENE_1311658
|
programmed cell death 1 |
| chr3_+_172550258 | 6.56 |
ENSRNOT00000075148
|
Tubb1
|
tubulin, beta 1 class VI |
| chr16_+_10417185 | 6.56 |
ENSRNOT00000082186
|
Anxa8
|
annexin A8 |
| chr1_-_82004538 | 6.45 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr10_-_85825735 | 6.39 |
ENSRNOT00000055379
|
Fbxo47
|
F-box protein 47 |
| chr5_+_122100099 | 6.17 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr2_-_91497091 | 6.13 |
ENSRNOT00000015185
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr12_-_51965779 | 6.10 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr10_+_72147816 | 5.96 |
ENSRNOT00000090085
|
LOC102553386
|
SUMO-conjugating enzyme UBC9-like |
| chrX_+_71335491 | 5.96 |
ENSRNOT00000076003
|
Nono
|
non-POU domain containing, octamer-binding |
| chr1_+_16478127 | 5.88 |
ENSRNOT00000019076
|
Ahi1
|
Abelson helper integration site 1 |
| chrX_+_14019961 | 5.72 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr6_-_140715174 | 5.55 |
ENSRNOT00000085345
|
AABR07065773.1
|
|
| chr10_-_66848388 | 5.54 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr7_-_104800536 | 5.51 |
ENSRNOT00000085650
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr16_-_32439421 | 5.49 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chr2_+_4989295 | 5.48 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr9_+_91001828 | 5.44 |
ENSRNOT00000080956
|
AABR07068214.1
|
|
| chr8_-_21492801 | 5.43 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr9_+_92681078 | 5.43 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr4_+_119842013 | 5.43 |
ENSRNOT00000057286
|
Hmces
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr2_+_143895982 | 5.32 |
ENSRNOT00000080026
|
Supt20h
|
SPT20 homolog, SAGA complex component |
| chr3_+_48626038 | 5.29 |
ENSRNOT00000009697
|
Gca
|
grancalcin |
| chr3_-_26056818 | 5.28 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr9_+_4421069 | 5.27 |
ENSRNOT00000065721
|
Sgo1
|
shugoshin 1 |
| chr2_-_39007976 | 5.20 |
ENSRNOT00000064292
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr2_-_88414012 | 5.17 |
ENSRNOT00000014762
|
Lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr1_-_201942344 | 5.15 |
ENSRNOT00000027956
|
RGD1305014
|
similar to RIKEN cDNA 2310057M21 |
| chr18_-_86279680 | 5.12 |
ENSRNOT00000006169
|
LOC689166
|
hypothetical protein LOC689166 |
| chr13_-_89874008 | 5.05 |
ENSRNOT00000051368
|
Ptma
|
prothymosin alpha |
| chr11_+_47146308 | 5.03 |
ENSRNOT00000002191
|
Cep97
|
centrosomal protein 97 |
| chr3_+_11554457 | 5.00 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr3_+_112950890 | 4.95 |
ENSRNOT00000015472
|
Ccndbp1
|
cyclin D1 binding protein 1 |
| chr5_-_62187930 | 4.93 |
ENSRNOT00000011787
|
Coro2a
|
coronin 2A |
| chr18_+_3163214 | 4.87 |
ENSRNOT00000017291
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr8_-_2045817 | 4.87 |
ENSRNOT00000009542
ENSRNOT00000081171 ENSRNOT00000078765 |
Gria4
|
glutamate ionotropic receptor AMPA type subunit 4 |
| chr8_-_55276070 | 4.70 |
ENSRNOT00000041555
|
LOC108351703
|
60S ribosomal protein L27a-like |
| chr1_-_20962526 | 4.70 |
ENSRNOT00000061332
ENSRNOT00000017322 ENSRNOT00000017412 ENSRNOT00000079688 ENSRNOT00000017417 |
Epb41l2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr2_-_165591110 | 4.64 |
ENSRNOT00000091140
|
Ift80
|
intraflagellar transport 80 |
| chr9_-_46309451 | 4.64 |
ENSRNOT00000018684
|
Rnf149
|
ring finger protein 149 |
| chr3_+_47677720 | 4.54 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr16_-_64778486 | 4.53 |
ENSRNOT00000031701
|
Rnf122
|
ring finger protein 122 |
| chr1_-_211956858 | 4.47 |
ENSRNOT00000054886
|
Cfap46
|
cilia and flagella associated protein 46 |
| chr5_-_155709215 | 4.35 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr8_-_96985558 | 4.33 |
ENSRNOT00000018553
|
LOC501033
|
similar to UPF0258 protein KIAA1024 |
| chr18_-_1828606 | 4.31 |
ENSRNOT00000033312
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr3_-_61494778 | 4.31 |
ENSRNOT00000068018
|
Lnpk
|
lunapark, ER junction formation factor |
| chrX_-_113228265 | 4.28 |
ENSRNOT00000060001
|
RGD1566136
|
similar to 40S ribosomal protein S9 |
| chr5_-_9094616 | 4.28 |
ENSRNOT00000072210
|
Sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr9_-_80295446 | 4.28 |
ENSRNOT00000023769
|
Tnp1
|
transition protein 1 |
| chr4_-_119591817 | 4.24 |
ENSRNOT00000048768
|
AC097129.1
|
|
| chr13_-_26769374 | 4.18 |
ENSRNOT00000003768
|
Bcl2
|
BCL2, apoptosis regulator |
| chr19_+_26022849 | 4.18 |
ENSRNOT00000014887
|
Dnase2
|
deoxyribonuclease 2, lysosomal |
| chr5_-_19559244 | 4.14 |
ENSRNOT00000014289
ENSRNOT00000089666 |
Nsmaf
|
neutral sphingomyelinase activation associated factor |
| chr2_-_184263564 | 4.12 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr10_-_103340922 | 4.07 |
ENSRNOT00000004191
|
Btbd17
|
BTB domain containing 17 |
| chr8_+_36766977 | 4.03 |
ENSRNOT00000017648
|
Pus3
|
pseudouridylate synthase 3 |
| chr3_+_160467552 | 3.99 |
ENSRNOT00000066657
|
Stk4
|
serine/threonine kinase 4 |
| chr14_+_81858737 | 3.89 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr14_-_44225713 | 3.88 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr2_-_207350812 | 3.87 |
ENSRNOT00000056015
|
Capza1
|
capping actin protein of muscle Z-line alpha subunit 1 |
| chr8_+_99636749 | 3.84 |
ENSRNOT00000086524
|
Plscr1
|
phospholipid scramblase 1 |
| chrX_-_15504165 | 3.83 |
ENSRNOT00000006233
|
Otud5
|
OTU deubiquitinase 5 |
| chr2_+_92559929 | 3.80 |
ENSRNOT00000033404
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr17_-_27452314 | 3.75 |
ENSRNOT00000018936
|
Riok1
|
RIO kinase 1 |
| chr2_+_243422811 | 3.75 |
ENSRNOT00000014694
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr11_+_64472072 | 3.75 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr18_+_86116044 | 3.74 |
ENSRNOT00000058160
ENSRNOT00000076159 |
Rttn
|
rotatin |
| chr17_-_42226377 | 3.69 |
ENSRNOT00000024536
|
RGD1307443
|
similar to mKIAA0319 protein |
| chr18_-_6782757 | 3.68 |
ENSRNOT00000068150
|
Aqp4
|
aquaporin 4 |
| chr5_+_144031402 | 3.63 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr4_-_98593664 | 3.59 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr1_-_169534896 | 3.58 |
ENSRNOT00000082645
|
Trim30c
|
tripartite motif-containing 30C |
| chr1_+_228690119 | 3.57 |
ENSRNOT00000087169
|
Pfpl
|
pore forming protein-like |
| chr15_+_80040842 | 3.49 |
ENSRNOT00000043065
|
RGD1306441
|
similar to RIKEN cDNA 4921530L21 |
| chr2_+_43271092 | 3.49 |
ENSRNOT00000017571
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr10_-_110701137 | 3.46 |
ENSRNOT00000074193
|
Znf750
|
zinc finger protein 750 |
| chr1_+_93242050 | 3.46 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr1_+_266781617 | 3.46 |
ENSRNOT00000027417
|
Ina
|
internexin neuronal intermediate filament protein, alpha |
| chr17_+_16242365 | 3.43 |
ENSRNOT00000022669
|
Phf2
|
PHD finger protein 2 |
| chr18_-_57515834 | 3.42 |
ENSRNOT00000026098
|
Adrb2
|
adrenoceptor beta 2 |
| chr18_-_74151950 | 3.40 |
ENSRNOT00000023101
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr4_+_110699557 | 3.38 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr3_-_105470475 | 3.35 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr11_+_74984613 | 3.35 |
ENSRNOT00000035049
|
Atp13a5
|
ATPase 13A5 |
| chr20_-_6195463 | 3.31 |
ENSRNOT00000092474
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr1_-_233140237 | 3.26 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr4_-_129430251 | 3.25 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr2_-_43067956 | 3.20 |
ENSRNOT00000090616
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr4_-_28437676 | 3.18 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr16_-_54137660 | 3.16 |
ENSRNOT00000085435
|
Pcm1
|
pericentriolar material 1 |
| chr11_-_46693565 | 3.16 |
ENSRNOT00000044900
|
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr10_-_56429748 | 3.11 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chrY_-_1238420 | 3.07 |
ENSRNOT00000092078
|
Ddx3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3 |
| chr2_-_128675408 | 3.06 |
ENSRNOT00000019256
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr1_+_212578450 | 3.05 |
ENSRNOT00000064501
|
Paox
|
polyamine oxidase |
| chr1_-_98570949 | 3.04 |
ENSRNOT00000033648
|
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr2_-_144808245 | 3.04 |
ENSRNOT00000087849
|
LOC102552324
|
uncharacterized LOC102552324 |
| chr3_+_149466770 | 3.02 |
ENSRNOT00000065104
|
Bpifa6
|
BPI fold containing family A, member 6 |
| chr9_+_94928489 | 3.00 |
ENSRNOT00000087366
ENSRNOT00000085770 ENSRNOT00000024733 |
Sag
|
S-antigen visual arrestin |
| chr14_+_33247274 | 2.97 |
ENSRNOT00000075226
|
Spink2
|
serine peptidase inhibitor, Kazal type 2 |
| chr18_-_6782996 | 2.87 |
ENSRNOT00000090320
|
Aqp4
|
aquaporin 4 |
| chr1_-_44615760 | 2.87 |
ENSRNOT00000022148
|
Nox3
|
NADPH oxidase 3 |
| chr2_-_244335165 | 2.85 |
ENSRNOT00000084860
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr5_+_173340060 | 2.83 |
ENSRNOT00000067841
|
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr12_-_21450204 | 2.82 |
ENSRNOT00000065866
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr16_-_54137485 | 2.81 |
ENSRNOT00000014202
|
Pcm1
|
pericentriolar material 1 |
| chr2_-_250923744 | 2.73 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr13_+_90116843 | 2.71 |
ENSRNOT00000006306
|
Cd48
|
Cd48 molecule |
| chr14_-_44375804 | 2.71 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chrX_+_6273733 | 2.69 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr7_+_42304534 | 2.66 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr4_-_120041238 | 2.65 |
ENSRNOT00000073799
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr1_-_245514252 | 2.64 |
ENSRNOT00000033724
|
Pum3
|
pumilio RNA-binding family member 3 |
| chr18_+_30023828 | 2.64 |
ENSRNOT00000079008
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_-_133232322 | 2.63 |
ENSRNOT00000034487
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr8_+_117126692 | 2.59 |
ENSRNOT00000083046
ENSRNOT00000085609 |
Usp4
|
ubiquitin specific peptidase 4 |
| chr15_-_29922522 | 2.59 |
ENSRNOT00000081778
|
AABR07017669.2
|
|
| chr7_+_27620458 | 2.57 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr2_+_71753816 | 2.57 |
ENSRNOT00000025809
|
AABR07008724.1
|
|
| chr8_-_6034142 | 2.52 |
ENSRNOT00000014560
|
Birc2
|
baculoviral IAP repeat-containing 2 |
| chr3_+_81134505 | 2.50 |
ENSRNOT00000067664
|
Phf21a
|
PHD finger protein 21A |
| chr14_+_88543630 | 2.35 |
ENSRNOT00000060515
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr8_-_114766803 | 2.33 |
ENSRNOT00000047563
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr1_+_101410019 | 2.27 |
ENSRNOT00000042039
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr1_+_102924059 | 2.26 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr6_-_98666007 | 2.24 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr1_-_262013619 | 2.22 |
ENSRNOT00000021278
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr10_-_90501819 | 2.22 |
ENSRNOT00000050474
|
Gpatch8
|
G patch domain containing 8 |
| chrX_-_135419704 | 2.21 |
ENSRNOT00000044435
|
Zfp280c
|
zinc finger protein 280C |
| chr10_+_14062331 | 2.16 |
ENSRNOT00000029652
|
Noxo1
|
NADPH oxidase organizer 1 |
| chr20_-_4390436 | 2.14 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr7_+_27089278 | 2.14 |
ENSRNOT00000082398
|
Nfyb
|
nuclear transcription factor Y subunit beta |
| chr3_-_3691972 | 2.13 |
ENSRNOT00000061735
|
Qsox2
|
quiescin sulfhydryl oxidase 2 |
| chr1_+_193844538 | 2.13 |
ENSRNOT00000065311
|
AC123018.1
|
|
| chr4_-_142970245 | 2.09 |
ENSRNOT00000067815
|
AC112018.1
|
|
| chrX_+_22313028 | 2.06 |
ENSRNOT00000082725
|
Kdm5c
|
lysine demethylase 5C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 24.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 6.7 | 33.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 4.6 | 13.9 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 4.6 | 27.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 3.9 | 15.4 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 3.5 | 10.6 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 3.5 | 10.5 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 2.9 | 20.0 | GO:0070269 | pyroptosis(GO:0070269) |
| 2.5 | 7.5 | GO:0035444 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 2.4 | 7.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 2.2 | 13.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 2.2 | 13.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 2.1 | 6.4 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 2.1 | 14.6 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 2.0 | 9.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 1.8 | 5.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.7 | 13.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 1.6 | 6.6 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 1.5 | 6.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 1.5 | 5.9 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 1.5 | 4.4 | GO:0060661 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 1.4 | 4.2 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 1.4 | 5.5 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 1.4 | 4.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 1.3 | 3.8 | GO:1905077 | regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 1.1 | 3.4 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 1.1 | 3.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) diaphragm contraction(GO:0002086) |
| 1.1 | 4.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 1.0 | 11.4 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 1.0 | 4.0 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 1.0 | 4.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.0 | 11.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.9 | 6.6 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.9 | 3.6 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.9 | 5.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.9 | 6.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.8 | 2.5 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.8 | 8.4 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.8 | 4.9 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.7 | 11.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.7 | 7.4 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.7 | 2.7 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.7 | 6.0 | GO:1903376 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.7 | 3.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.6 | 4.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.6 | 4.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.6 | 3.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.6 | 4.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.5 | 3.8 | GO:2000371 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.5 | 29.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.5 | 4.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.5 | 11.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 2.7 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 1.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 | 5.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.4 | 2.9 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.4 | 5.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.4 | 17.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.4 | 2.6 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.4 | 3.8 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.4 | 11.9 | GO:0051482 | positive regulation of Rho protein signal transduction(GO:0035025) positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.4 | 2.9 | GO:0048840 | otolith development(GO:0048840) |
| 0.4 | 1.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.3 | 3.5 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 1.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.3 | 3.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 1.7 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.3 | 26.7 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.3 | 9.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 5.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 4.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.3 | 1.5 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.3 | 2.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 2.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 3.0 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.3 | 5.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.3 | 3.4 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.3 | 1.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 0.5 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.2 | 4.9 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.2 | 6.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 3.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 4.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.5 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 2.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 12.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 2.5 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.2 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 5.5 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.2 | 2.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 8.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.2 | 1.9 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 9.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 3.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.4 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 6.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 4.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.4 | GO:1903925 | response to bisphenol A(GO:1903925) |
| 0.1 | 2.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 3.7 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 5.2 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 2.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 5.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 6.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 2.3 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) |
| 0.1 | 3.3 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 19.8 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.1 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 8.9 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 3.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.1 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 1.2 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 4.6 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 1.2 | GO:0007567 | parturition(GO:0007567) |
| 0.1 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.6 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) peptidyl-arginine modification(GO:0018195) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 4.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 3.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 5.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.9 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 1.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.1 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 4.0 | 20.0 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 2.5 | 7.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 2.5 | 9.8 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.1 | 3.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.9 | 6.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.8 | 4.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.8 | 4.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.8 | 5.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.8 | 5.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.7 | 13.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.6 | 3.9 | GO:0071203 | WASH complex(GO:0071203) |
| 0.6 | 23.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.5 | 5.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 11.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 2.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.4 | 5.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.4 | 3.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 15.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 3.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 3.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 19.5 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.3 | 17.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.6 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.3 | 25.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 2.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 11.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 1.5 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 1.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 24.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 2.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 22.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 5.3 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.2 | 4.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 3.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 3.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 2.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 13.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 43.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 9.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 5.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0044317 | rod spherule(GO:0044317) |
| 0.1 | 12.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 8.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 5.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 11.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 3.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 4.9 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 3.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 22.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 2.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 3.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 4.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 10.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 14.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 7.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 6.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 4.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 5.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 6.0 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 24.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 4.8 | 14.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 4.6 | 13.9 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 2.5 | 7.5 | GO:0015099 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) chromium ion transmembrane transporter activity(GO:0070835) |
| 1.6 | 13.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.6 | 26.0 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 1.5 | 10.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 1.5 | 4.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 1.4 | 8.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 1.4 | 4.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 1.4 | 9.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 1.2 | 3.6 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.1 | 3.4 | GO:0031711 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 1.1 | 7.4 | GO:0043426 | MRF binding(GO:0043426) |
| 1.0 | 3.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 1.0 | 7.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.0 | 4.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.9 | 4.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.9 | 18.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.9 | 14.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.9 | 33.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.8 | 4.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.8 | 3.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.8 | 3.8 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.7 | 2.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.7 | 4.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.7 | 6.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.7 | 5.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.6 | 2.6 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.6 | 3.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.6 | 11.4 | GO:0001618 | virus receptor activity(GO:0001618) mannose binding(GO:0005537) |
| 0.5 | 25.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.5 | 6.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 12.3 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.4 | 4.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 7.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 2.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.4 | 3.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.3 | 3.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 1.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.3 | 6.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 2.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 6.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.3 | 7.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 4.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 6.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 4.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.3 | 1.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-dependent protein binding(GO:0032767) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 2.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 1.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 3.9 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.2 | 25.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 2.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.2 | 4.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.2 | 5.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 21.5 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.2 | 27.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 5.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 3.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 19.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 1.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 8.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 2.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 3.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 7.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 8.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 4.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 2.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.4 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 5.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 6.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 1.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 3.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 8.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 1.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 5.3 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 4.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 3.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 21.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 4.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.5 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0003684 | p53 binding(GO:0002039) damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 7.1 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 18.5 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 3.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 24.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.9 | 13.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.6 | 8.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 5.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 9.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 19.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.4 | 4.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 6.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 5.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 12.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 7.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 13.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 17.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 5.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 29.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.5 | 14.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.3 | 24.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 1.2 | 9.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.0 | 18.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.9 | 10.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.8 | 19.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.7 | 10.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.7 | 8.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.5 | 6.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 15.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.4 | 13.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 14.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.4 | 9.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.4 | 8.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 18.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 57.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.3 | 3.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 8.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 7.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 4.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 4.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 4.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 4.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 2.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 9.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 3.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 7.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 4.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 5.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 19.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 11.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 6.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.5 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 8.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |