Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Nkx2-1
Z-value: 0.38
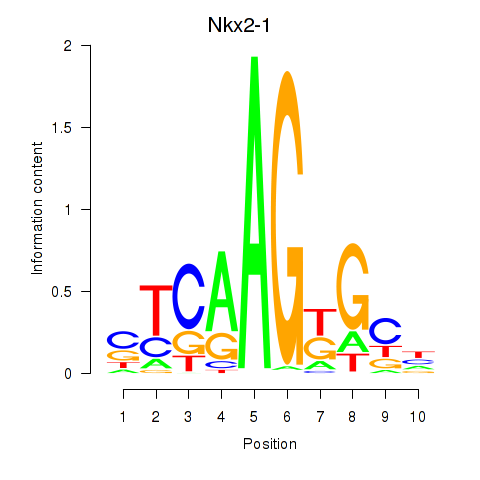
Transcription factors associated with Nkx2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-1
|
ENSRNOG00000008644 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-1 | rn6_v1_chr6_-_77421286_77421286 | 0.21 | 1.8e-04 | Click! |
Activity profile of Nkx2-1 motif
Sorted Z-values of Nkx2-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_100166863 | 12.44 |
ENSRNOT00000014505
|
Sftpb
|
surfactant protein B |
| chr10_+_11240138 | 11.91 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr11_+_66713888 | 8.81 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr10_+_40247436 | 7.95 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr13_-_82005741 | 6.91 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr1_+_33910912 | 5.15 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr5_-_22769907 | 4.66 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr2_+_202200797 | 4.11 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr12_+_41486076 | 3.52 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr5_+_2353468 | 3.52 |
ENSRNOT00000090346
|
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr1_-_207811008 | 3.38 |
ENSRNOT00000080506
|
Clrn3
|
clarin 3 |
| chr3_-_168018410 | 3.11 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr1_+_222519615 | 2.92 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr1_-_207766250 | 2.53 |
ENSRNOT00000031397
|
Clrn3
|
clarin 3 |
| chr1_-_141470380 | 2.33 |
ENSRNOT00000065759
|
Plin1
|
perilipin 1 |
| chr2_-_193101051 | 2.27 |
ENSRNOT00000065242
|
AABR07012318.1
|
|
| chr2_-_193063357 | 2.19 |
ENSRNOT00000072995
|
AABR07012317.2
|
|
| chr16_+_86631064 | 2.00 |
ENSRNOT00000019675
|
Efnb2
|
ephrin B2 |
| chr10_-_62664466 | 1.99 |
ENSRNOT00000078730
|
Git1
|
GIT ArfGAP 1 |
| chr10_+_89069256 | 1.96 |
ENSRNOT00000008576
|
Tubg2
|
tubulin, gamma 2 |
| chr1_+_86949413 | 1.81 |
ENSRNOT00000064153
|
Sirt2
|
sirtuin 2 |
| chr10_+_16635989 | 1.63 |
ENSRNOT00000028155
|
Nkx2-5
|
NK2 homeobox 5 |
| chr2_-_250232295 | 1.49 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr6_-_98666007 | 1.35 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr4_-_145454834 | 1.24 |
ENSRNOT00000013154
ENSRNOT00000056493 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr18_+_30172740 | 1.21 |
ENSRNOT00000027340
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_+_243589607 | 1.13 |
ENSRNOT00000021792
|
Dmrt3
|
doublesex and mab-3 related transcription factor 3 |
| chr10_-_90393317 | 1.12 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chrX_+_136460215 | 1.10 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr2_+_188844073 | 1.06 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr2_-_188553289 | 0.95 |
ENSRNOT00000088822
|
Trim46
|
tripartite motif-containing 46 |
| chr4_-_165787668 | 0.83 |
ENSRNOT00000044029
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr17_-_43640387 | 0.71 |
ENSRNOT00000087731
|
Hist1h1c
|
histone cluster 1 H1 family member c |
| chr6_+_128973956 | 0.71 |
ENSRNOT00000075399
|
Fam181a
|
family with sequence similarity 181, member A |
| chr6_+_101924000 | 0.70 |
ENSRNOT00000092157
|
Mpp5
|
membrane palmitoylated protein 5 |
| chr11_-_78456200 | 0.64 |
ENSRNOT00000067251
ENSRNOT00000036179 |
Tp63
|
tumor protein p63 |
| chr5_-_145423172 | 0.60 |
ENSRNOT00000081919
|
Gjb5
|
gap junction protein, beta 5 |
| chr7_+_28654733 | 0.54 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr7_+_107467260 | 0.48 |
ENSRNOT00000009240
ENSRNOT00000077382 |
Tg
|
thyroglobulin |
| chr6_+_101923785 | 0.44 |
ENSRNOT00000012005
|
Mpp5
|
membrane palmitoylated protein 5 |
| chr1_-_40921508 | 0.43 |
ENSRNOT00000026433
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr10_-_90096096 | 0.43 |
ENSRNOT00000081539
|
Tmem101
|
transmembrane protein 101 |
| chr20_-_4391402 | 0.41 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_78456501 | 0.40 |
ENSRNOT00000036193
|
Tp63
|
tumor protein p63 |
| chr7_+_116745061 | 0.37 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr1_-_170694872 | 0.37 |
ENSRNOT00000075443
|
LOC100911839
|
olfactory receptor 2D3-like |
| chr1_-_227441442 | 0.37 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr7_+_16304954 | 0.36 |
ENSRNOT00000050751
|
Olr927
|
olfactory receptor 927 |
| chr5_-_127273656 | 0.36 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chrX_+_86126157 | 0.31 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr7_-_69982592 | 0.31 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr18_-_75207306 | 0.30 |
ENSRNOT00000021717
|
Setbp1
|
SET binding protein 1 |
| chr6_+_27241919 | 0.28 |
ENSRNOT00000013123
|
Cib4
|
calcium and integrin binding family member 4 |
| chr1_-_255976425 | 0.25 |
ENSRNOT00000085303
|
Ide
|
insulin degrading enzyme |
| chr13_+_47935163 | 0.20 |
ENSRNOT00000006768
|
Eif2d
|
eukaryotic translation initiation factor 2D |
| chr9_+_10061978 | 0.19 |
ENSRNOT00000075180
|
Acer1
|
alkaline ceramidase 1 |
| chr1_-_233140237 | 0.17 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr6_+_115170865 | 0.16 |
ENSRNOT00000005671
|
Tshr
|
thyroid stimulating hormone receptor |
| chr4_-_71080499 | 0.14 |
ENSRNOT00000074623
|
RGD1562066
|
similar to odorant receptor ORZ6 |
| chr10_-_84789832 | 0.14 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr17_+_1305016 | 0.13 |
ENSRNOT00000025965
|
Ercc6l2
|
ERCC excision repair 6 like 2 |
| chrX_+_145558840 | 0.11 |
ENSRNOT00000032522
|
4931400O07Rik
|
RIKEN cDNA 4931400O07 gene |
| chr16_+_49185560 | 0.06 |
ENSRNOT00000014360
|
Helt
|
helt bHLH transcription factor |
| chr15_-_51386190 | 0.05 |
ENSRNOT00000022706
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 11.9 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 2.5 | 12.4 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 2.3 | 6.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 1.3 | 5.2 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 1.1 | 7.9 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 1.0 | 4.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 4.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.9 | 3.5 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.7 | 2.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.6 | 1.8 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.5 | 1.6 | GO:0003285 | atrioventricular node development(GO:0003162) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) septum secundum development(GO:0003285) His-Purkinje system cell differentiation(GO:0060932) |
| 0.3 | 1.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 3.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 2.0 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 1.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 2.0 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.5 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 2.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.9 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.2 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.2 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 8.3 | GO:0042692 | muscle cell differentiation(GO:0042692) |
| 0.0 | 1.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 12.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.8 | 4.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.4 | 4.7 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 3.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 0.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 3.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 1.0 | 7.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 1.8 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 1.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 3.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.0 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 13.0 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 8.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 11.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.6 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 2.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |