Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
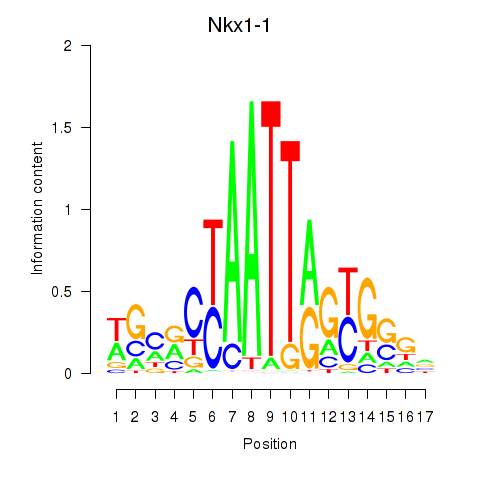
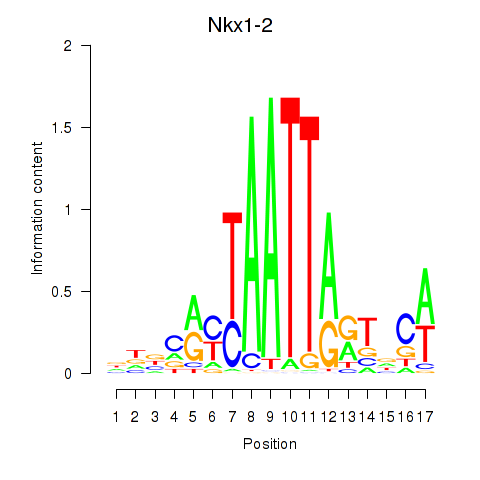
Results for Nkx1-1_Nkx1-2
Z-value: 0.40
Transcription factors associated with Nkx1-1_Nkx1-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx1-1
|
ENSRNOG00000005113 | NK1 homeobox 1 |
|
Nkx1-2
|
ENSRNOG00000017006 | NK1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx1-2 | rn6_v1_chr1_-_204605552_204605552 | -0.32 | 3.8e-09 | Click! |
| Nkx1-1 | rn6_v1_chr14_+_82603671_82603671 | -0.08 | 1.4e-01 | Click! |
Activity profile of Nkx1-1_Nkx1-2 motif
Sorted Z-values of Nkx1-1_Nkx1-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_168734296 | 19.09 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chrX_+_14019961 | 13.67 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr12_-_35979193 | 11.82 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr9_+_73418607 | 9.60 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr2_+_145174876 | 9.28 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chrX_+_15113878 | 8.52 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr4_-_168656673 | 7.50 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr1_+_278557792 | 7.15 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr7_-_76488216 | 6.78 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chrX_+_106360393 | 6.31 |
ENSRNOT00000004260
|
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chr19_+_45938915 | 6.29 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr7_+_2752680 | 5.91 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr8_-_84506328 | 5.79 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr1_+_59156251 | 5.62 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr4_-_157304653 | 5.34 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr9_+_20241062 | 5.17 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr3_-_120076788 | 5.02 |
ENSRNOT00000047158
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr16_-_3765917 | 4.80 |
ENSRNOT00000088284
|
Duxbl1
|
double homeobox B-like 1 |
| chr17_+_63635086 | 4.66 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr12_-_46493203 | 4.53 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr12_+_49665282 | 4.27 |
ENSRNOT00000086397
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr4_-_55011415 | 4.23 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr17_-_52477575 | 4.15 |
ENSRNOT00000081290
|
Gli3
|
GLI family zinc finger 3 |
| chr5_+_71742911 | 3.98 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr1_-_261179790 | 3.88 |
ENSRNOT00000074420
ENSRNOT00000072073 |
Exosc1
|
exosome component 1 |
| chr5_-_161981441 | 3.87 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr6_+_111076351 | 3.66 |
ENSRNOT00000089644
|
Tmem63c
|
transmembrane protein 63c |
| chr17_-_87826421 | 3.60 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr9_+_71915421 | 3.35 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr1_-_100926335 | 3.27 |
ENSRNOT00000020304
|
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr15_+_43582020 | 3.25 |
ENSRNOT00000013001
|
Pnma2
|
paraneoplastic Ma antigen 2 |
| chr13_+_92569785 | 3.20 |
ENSRNOT00000082700
|
Fmn2
|
formin 2 |
| chr8_+_22648323 | 3.10 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr1_+_53360157 | 2.99 |
ENSRNOT00000017809
|
Rps6ka2
|
ribosomal protein S6 kinase A2 |
| chr6_-_108660063 | 2.82 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr2_-_149432106 | 2.77 |
ENSRNOT00000051027
|
P2ry13
|
purinergic receptor P2Y13 |
| chr18_-_16543992 | 2.74 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr9_-_30844199 | 2.73 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr17_-_14627937 | 2.64 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr2_-_196177775 | 2.59 |
ENSRNOT00000028553
|
Zfp687
|
zinc finger protein 687 |
| chr5_+_58995249 | 2.58 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr1_+_101687855 | 2.47 |
ENSRNOT00000028546
ENSRNOT00000091597 |
Dbp
|
D-box binding PAR bZIP transcription factor |
| chr8_-_21492801 | 2.46 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr10_+_29289203 | 2.45 |
ENSRNOT00000067013
|
Pwwp2a
|
PWWP domain containing 2A |
| chr12_-_12738620 | 2.41 |
ENSRNOT00000078603
ENSRNOT00000067021 |
Pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr16_+_60307148 | 2.30 |
ENSRNOT00000015239
|
Eri1
|
exoribonuclease 1 |
| chr1_-_84008293 | 2.28 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr4_+_158088505 | 2.28 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr10_+_74413989 | 2.28 |
ENSRNOT00000036098
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_131694755 | 2.23 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr18_-_69944632 | 2.13 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr5_+_131380297 | 2.12 |
ENSRNOT00000010548
|
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr1_+_220869805 | 2.11 |
ENSRNOT00000015962
|
Cfl1
|
cofilin 1 |
| chr1_+_15834779 | 2.10 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr3_+_62648447 | 2.10 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr10_+_92628356 | 2.08 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr10_+_48569555 | 2.08 |
ENSRNOT00000003966
|
Adora2b
|
adenosine A2B receptor |
| chr16_-_10802512 | 1.96 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr20_+_6211420 | 1.96 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr1_-_215284548 | 1.95 |
ENSRNOT00000090375
|
Cttn
|
cortactin |
| chr8_-_78096302 | 1.91 |
ENSRNOT00000086185
ENSRNOT00000077999 |
Myzap
|
myocardial zonula adherens protein |
| chr8_-_33121002 | 1.89 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr7_+_35773928 | 1.86 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr8_+_42663633 | 1.84 |
ENSRNOT00000048537
|
Olr1266
|
olfactory receptor 1266 |
| chr1_+_141767940 | 1.80 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr11_+_31389514 | 1.77 |
ENSRNOT00000000325
|
Olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr13_+_67611708 | 1.67 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr5_+_107369730 | 1.65 |
ENSRNOT00000046837
|
LOC100912314
|
interferon alpha-1-like |
| chr17_-_78812111 | 1.65 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr1_+_7252349 | 1.61 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr6_-_44361908 | 1.57 |
ENSRNOT00000009491
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr1_+_229416489 | 1.46 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chr1_-_64147251 | 1.45 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chrX_+_73390903 | 1.40 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chr12_+_12738812 | 1.39 |
ENSRNOT00000092233
ENSRNOT00000001379 |
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr15_-_80713153 | 1.32 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr3_+_14889510 | 1.28 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr13_-_50916982 | 1.23 |
ENSRNOT00000004408
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr1_+_211582077 | 1.21 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr14_-_34528262 | 1.11 |
ENSRNOT00000003010
|
Tmem165
|
transmembrane protein 165 |
| chr6_+_18880737 | 1.11 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_-_33025271 | 1.10 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_+_106393959 | 1.07 |
ENSRNOT00000092168
|
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr20_+_12944786 | 1.04 |
ENSRNOT00000048218
|
Pcnt
|
pericentrin |
| chr5_-_107447636 | 1.02 |
ENSRNOT00000040254
|
Ifna16l1
|
interferon, alpha 16-like 1 |
| chr14_-_113644779 | 1.02 |
ENSRNOT00000005306
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr1_+_224998172 | 0.97 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr14_+_84465515 | 0.91 |
ENSRNOT00000035591
|
Osm
|
oncostatin M |
| chr16_+_7035241 | 0.89 |
ENSRNOT00000084977
|
Nek4
|
NIMA-related kinase 4 |
| chr3_-_124879216 | 0.83 |
ENSRNOT00000028886
|
Tmem230
|
transmembrane protein 230 |
| chr1_-_173393390 | 0.78 |
ENSRNOT00000050657
|
Olr285
|
olfactory receptor 285 |
| chr5_-_154222702 | 0.77 |
ENSRNOT00000080069
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr5_+_165405168 | 0.77 |
ENSRNOT00000014934
|
Srm
|
spermidine synthase |
| chr1_-_216663720 | 0.76 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr3_+_80081647 | 0.73 |
ENSRNOT00000057161
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr5_+_36566783 | 0.72 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr7_+_71157664 | 0.71 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr10_+_87759769 | 0.70 |
ENSRNOT00000017378
ENSRNOT00000046526 |
Krtap9-1
|
keratin associated protein 9-1 |
| chr16_+_39145230 | 0.65 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr7_+_3246220 | 0.60 |
ENSRNOT00000009155
|
Sarnp
|
SAP domain containing ribonucleoprotein |
| chrX_+_111396995 | 0.60 |
ENSRNOT00000087634
|
Pih1d3
|
PIH1 domain containing 3 |
| chr3_-_90751055 | 0.59 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr13_-_35933243 | 0.53 |
ENSRNOT00000039074
|
Tmem177
|
transmembrane protein 177 |
| chr11_-_51202703 | 0.50 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr10_-_65502936 | 0.49 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr5_+_144114012 | 0.48 |
ENSRNOT00000035597
|
Stk40
|
serine/threonine kinase 40 |
| chr18_+_16544508 | 0.46 |
ENSRNOT00000020601
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr8_-_43796224 | 0.45 |
ENSRNOT00000084315
|
Olr1330
|
olfactory receptor 1330 |
| chr1_-_89007041 | 0.41 |
ENSRNOT00000032363
|
Proser3
|
proline and serine rich 3 |
| chr3_+_16241573 | 0.41 |
ENSRNOT00000082831
|
Olr407
|
olfactory receptor 407 |
| chr1_-_101123402 | 0.40 |
ENSRNOT00000027976
|
Rpl13a
|
|
| chr5_+_154311998 | 0.38 |
ENSRNOT00000084085
|
Gale
|
UDP-galactose-4-epimerase |
| chr16_+_39353283 | 0.37 |
ENSRNOT00000080125
|
LOC108348202
|
disintegrin and metalloproteinase domain-containing protein 21-like |
| chr6_+_137323713 | 0.35 |
ENSRNOT00000029017
|
Pld4
|
phospholipase D family, member 4 |
| chr1_+_252284464 | 0.32 |
ENSRNOT00000027969
|
Lipf
|
lipase F, gastric type |
| chr5_-_106865746 | 0.30 |
ENSRNOT00000008224
|
Ifnb1
|
interferon beta 1 |
| chrX_+_140878216 | 0.30 |
ENSRNOT00000075840
ENSRNOT00000077026 |
Zic3
|
Zic family member 3 |
| chr6_-_59950586 | 0.28 |
ENSRNOT00000005800
|
Arl4a
|
ADP-ribosylation factor like GTPase 4A |
| chr8_-_19333047 | 0.19 |
ENSRNOT00000048500
|
Olr1144
|
olfactory receptor 1144 |
| chr14_+_81858737 | 0.16 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr10_-_31052102 | 0.16 |
ENSRNOT00000007917
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr5_-_154223536 | 0.16 |
ENSRNOT00000012258
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr2_+_248398917 | 0.13 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr1_-_169770188 | 0.10 |
ENSRNOT00000072602
|
Olr165
|
olfactory receptor 165 |
| chr14_+_36216002 | 0.09 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr2_-_219458271 | 0.08 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr16_+_7035068 | 0.07 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chr4_-_170740274 | 0.05 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx1-1_Nkx1-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0019086 | late viral transcription(GO:0019086) |
| 1.4 | 4.3 | GO:0042441 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 1.1 | 3.3 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 1.1 | 3.2 | GO:0070649 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 1.0 | 4.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 1.0 | 3.1 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.9 | 3.6 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.7 | 2.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.7 | 2.1 | GO:2000771 | regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) |
| 0.7 | 2.0 | GO:0048372 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) |
| 0.6 | 6.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.6 | 1.8 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.6 | 8.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.6 | 1.7 | GO:0045799 | MAPK import into nucleus(GO:0000189) regulation of chromatin assembly(GO:0010847) positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.5 | 1.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.5 | 3.9 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.5 | 1.4 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.4 | 3.0 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.4 | 4.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.4 | 1.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.4 | 1.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.4 | 2.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.3 | 2.1 | GO:0010701 | positive regulation of chronic inflammatory response(GO:0002678) positive regulation of norepinephrine secretion(GO:0010701) |
| 0.3 | 2.3 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.3 | 2.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 2.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 2.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 3.0 | GO:0033141 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.2 | 1.9 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.2 | 9.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 19.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.2 | 1.9 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 2.8 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.9 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 5.8 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 1.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 11.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 4.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 5.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 2.4 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 4.5 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 5.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 0.8 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 2.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 0.9 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 3.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 0.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 1.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 2.1 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 1.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 1.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 7.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 4.0 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 1.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 1.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 2.6 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 4.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 2.3 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.4 | GO:1904627 | cellular response to UV-B(GO:0071493) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.5 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 1.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 2.1 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 2.7 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 2.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 5.7 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 2.5 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 3.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.5 | 8.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.5 | 2.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.4 | 1.3 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.4 | 2.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 3.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 1.9 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.3 | 1.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 3.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 2.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 4.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.2 | 3.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 1.6 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 1.7 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 2.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 1.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 4.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 14.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 4.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 5.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 4.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.1 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 1.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 2.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 2.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 17.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 4.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 4.5 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.5 | GO:0005819 | spindle(GO:0005819) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.1 | 4.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.7 | 3.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.7 | 4.3 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.6 | 2.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.6 | 2.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.5 | 3.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.4 | 3.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 9.6 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.4 | 3.1 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.3 | 2.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 3.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 2.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 2.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 8.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 6.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 1.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 6.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 11.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 0.5 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.1 | 1.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 4.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.9 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.1 | 2.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 5.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 3.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 5.8 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.1 | 2.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 18.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 4.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 1.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 3.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 1.4 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 4.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 3.7 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 4.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 3.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 9.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 6.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 3.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 4.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 5.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 3.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 3.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 4.9 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 6.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 4.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 2.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 2.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |