Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
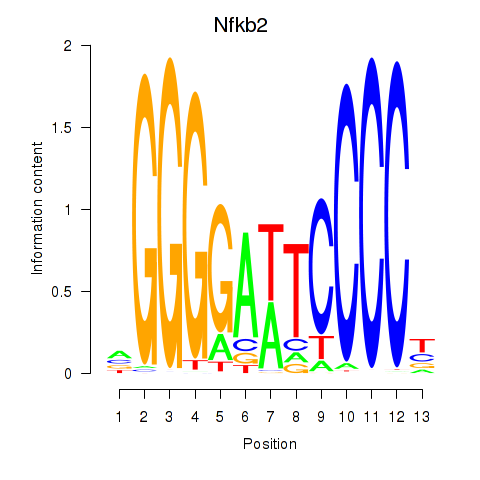
Results for Nfkb2
Z-value: 0.88
Transcription factors associated with Nfkb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfkb2
|
ENSRNOG00000019311 | nuclear factor kappa B subunit 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb2 | rn6_v1_chr1_+_266053002_266053002 | 0.77 | 4.8e-65 | Click! |
Activity profile of Nfkb2 motif
Sorted Z-values of Nfkb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_18315320 | 35.93 |
ENSRNOT00000006954
|
AABR07051626.2
|
|
| chr4_+_69384145 | 31.34 |
ENSRNOT00000084834
|
Trbv13-2
|
T cell receptor beta, variable 13-2 |
| chr3_+_18706988 | 30.54 |
ENSRNOT00000074650
|
AABR07051652.1
|
|
| chr20_-_4070721 | 30.31 |
ENSRNOT00000000523
|
RT1-Ba
|
RT1 class II, locus Ba |
| chr1_-_100537377 | 30.08 |
ENSRNOT00000026599
|
Spib
|
Spi-B transcription factor |
| chr2_+_223121410 | 28.10 |
ENSRNOT00000087559
|
AABR07013116.1
|
|
| chr3_-_20695952 | 25.45 |
ENSRNOT00000072306
|
AABR07051746.1
|
|
| chr20_-_4921348 | 22.87 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr1_-_89269930 | 22.20 |
ENSRNOT00000028532
|
Ffar2
|
free fatty acid receptor 2 |
| chr20_+_5414448 | 21.52 |
ENSRNOT00000078972
ENSRNOT00000080900 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr5_-_143120165 | 21.08 |
ENSRNOT00000012314
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr14_-_18839420 | 18.75 |
ENSRNOT00000034090
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr1_-_167347490 | 16.64 |
ENSRNOT00000076499
|
Rhog
|
ras homolog family member G |
| chr20_-_3397039 | 14.67 |
ENSRNOT00000001084
ENSRNOT00000085259 |
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr1_-_80544825 | 14.34 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr20_+_4852671 | 13.96 |
ENSRNOT00000001111
|
Lta
|
lymphotoxin alpha |
| chr20_+_5374985 | 13.49 |
ENSRNOT00000052270
|
RT1-A2
|
RT1 class Ia, locus A2 |
| chr13_+_47539231 | 13.44 |
ENSRNOT00000029694
|
Fcamr
|
Fc fragment of IgA and IgM receptor |
| chr6_+_132702448 | 13.19 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr4_-_115157263 | 13.18 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr7_-_143852119 | 13.04 |
ENSRNOT00000016801
|
Rarg
|
retinoic acid receptor, gamma |
| chr15_+_42489377 | 12.84 |
ENSRNOT00000064306
|
Pbk
|
PDZ binding kinase |
| chr20_+_4852496 | 12.01 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chr10_-_56412544 | 11.65 |
ENSRNOT00000020578
|
Tmem102
|
transmembrane protein 102 |
| chr7_+_133856101 | 10.87 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr4_+_171250818 | 10.81 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr20_-_3793985 | 10.63 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr7_+_70364813 | 10.31 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr15_+_62406873 | 10.11 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr13_+_111870121 | 10.02 |
ENSRNOT00000007333
|
Irf6
|
interferon regulatory factor 6 |
| chr6_-_41039437 | 9.56 |
ENSRNOT00000005774
|
Trib2
|
tribbles pseudokinase 2 |
| chr1_-_166413564 | 9.51 |
ENSRNOT00000026286
|
Atg16l2
|
autophagy related 16-like 2 |
| chr14_-_18745457 | 9.21 |
ENSRNOT00000003778
|
Cxcl1
|
C-X-C motif chemokine ligand 1 |
| chr2_-_31826867 | 8.76 |
ENSRNOT00000025687
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr1_+_221710670 | 8.21 |
ENSRNOT00000064798
|
Map4k2
|
mitogen activated protein kinase kinase kinase kinase 2 |
| chr2_+_189169877 | 8.19 |
ENSRNOT00000028225
|
She
|
Src homology 2 domain containing E |
| chr8_-_119326938 | 7.75 |
ENSRNOT00000044467
|
Ccrl2
|
C-C motif chemokine receptor like 2 |
| chr1_-_101046208 | 6.93 |
ENSRNOT00000091711
|
Prr12
|
proline rich 12 |
| chr1_-_170578693 | 6.88 |
ENSRNOT00000025651
|
Rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr15_-_28044210 | 6.46 |
ENSRNOT00000033809
|
Rnase1l1
|
ribonuclease, RNase A family, 1-like 1 (pancreatic) |
| chr1_+_170578889 | 6.09 |
ENSRNOT00000025906
|
Ilk
|
integrin-linked kinase |
| chr10_+_86514830 | 5.80 |
ENSRNOT00000048647
ENSRNOT00000009535 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr20_+_44521279 | 5.74 |
ENSRNOT00000085987
|
Fyn
|
FYN proto-oncogene, Src family tyrosine kinase |
| chr1_-_82580007 | 5.60 |
ENSRNOT00000078668
|
Axl
|
Axl receptor tyrosine kinase |
| chr1_-_266086299 | 5.07 |
ENSRNOT00000026609
|
Cuedc2
|
CUE domain containing 2 |
| chr1_+_189549960 | 5.04 |
ENSRNOT00000019654
|
Exnef
|
exonuclease NEF-sp |
| chr9_+_71230108 | 4.93 |
ENSRNOT00000018326
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr6_+_52663112 | 4.68 |
ENSRNOT00000013842
|
Atxn7l1
|
ataxin 7-like 1 |
| chr5_+_142986526 | 4.61 |
ENSRNOT00000012811
|
Rspo1
|
R-spondin 1 |
| chr10_+_75055020 | 4.19 |
ENSRNOT00000010755
|
Tspoap1
|
TSPO associated protein 1 |
| chr1_-_127599257 | 4.06 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr3_-_105214989 | 3.90 |
ENSRNOT00000037895
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr13_+_49870976 | 3.86 |
ENSRNOT00000090170
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr14_-_18839595 | 3.79 |
ENSRNOT00000078746
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr7_-_143217535 | 2.52 |
ENSRNOT00000088316
|
Kb15
|
type II keratin Kb15 |
| chr10_-_72188164 | 2.22 |
ENSRNOT00000085728
ENSRNOT00000042506 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr20_-_3299580 | 1.75 |
ENSRNOT00000050373
|
Gnl1
|
G protein nucleolar 1 |
| chr2_+_158097843 | 1.69 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr10_-_65200109 | 1.60 |
ENSRNOT00000030501
|
Nufip2
|
NUFIP2, FMR1 interacting protein 2 |
| chr12_-_38638536 | 1.54 |
ENSRNOT00000001690
|
Mlxip
|
MLX interacting protein |
| chrX_-_68562301 | 1.33 |
ENSRNOT00000076720
|
Ophn1
|
oligophrenin 1 |
| chr2_+_188748359 | 1.12 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr13_+_50873605 | 0.89 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr5_-_146973932 | 0.87 |
ENSRNOT00000007761
|
Zfp362
|
zinc finger protein 362 |
| chr10_-_74769637 | 0.82 |
ENSRNOT00000008889
|
LOC102555183
|
zinc finger protein OZF-like |
| chr2_+_140708397 | 0.61 |
ENSRNOT00000088846
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr4_-_77510202 | 0.46 |
ENSRNOT00000038550
|
Zfp786
|
zinc finger protein 786 |
| chr3_-_112084144 | 0.42 |
ENSRNOT00000010959
|
Pla2g4f
|
phospholipase A2, group IVF |
| chr20_-_3299420 | 0.35 |
ENSRNOT00000090999
|
Gnl1
|
G protein nucleolar 1 |
| chr10_-_44659707 | 0.09 |
ENSRNOT00000002064
|
RGD1559534
|
similar to Alpha enolase (2-phospho-D-glycerate hydro-lyase) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfkb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 26.0 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 7.2 | 21.5 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 7.0 | 21.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 5.6 | 22.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 4.3 | 13.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 3.3 | 13.2 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 3.1 | 9.2 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 2.4 | 9.6 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 2.0 | 7.9 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 1.8 | 10.8 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 1.6 | 4.9 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 1.6 | 30.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 1.6 | 9.5 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 1.4 | 5.6 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 1.3 | 13.5 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.2 | 11.6 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 1.0 | 10.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 1.0 | 6.9 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 1.0 | 3.9 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.0 | 5.7 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 1.0 | 14.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.9 | 8.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.9 | 10.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.8 | 5.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.8 | 22.5 | GO:1902624 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.8 | 30.1 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.7 | 22.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.5 | 16.6 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.5 | 8.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 4.6 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.5 | 10.0 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.4 | 1.7 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.3 | 5.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.3 | 12.8 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.2 | 3.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 6.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 7.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 2.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 6.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 2.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 1.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.4 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 1.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.9 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 57.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 3.0 | 21.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 2.9 | 8.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 1.9 | 30.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.6 | 4.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 1.6 | 14.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.5 | 13.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.4 | 6.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.8 | 13.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.6 | 10.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.4 | 5.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.4 | 5.6 | GO:0033643 | host cell part(GO:0033643) |
| 0.3 | 9.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 3.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 10.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 6.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 5.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 16.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 41.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 11.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 5.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 3.1 | 21.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 2.7 | 13.4 | GO:0019862 | IgA binding(GO:0019862) |
| 2.3 | 31.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.2 | 6.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.9 | 21.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 1.6 | 4.9 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 1.5 | 8.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.4 | 5.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.4 | 4.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 1.4 | 8.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 1.2 | 43.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.1 | 13.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.8 | 3.9 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.7 | 13.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.6 | 25.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.5 | 22.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.5 | 5.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 9.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.5 | 3.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 10.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 8.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.3 | 7.8 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.3 | 1.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 1.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 10.3 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 6.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 14.3 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 14.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 5.0 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 6.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 4.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 10.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 17.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 4.9 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 35.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.5 | 27.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.4 | 16.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.4 | 13.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.4 | 13.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 14.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.3 | 9.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.3 | 14.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 13.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 8.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.7 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 12.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 31.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.6 | 31.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 6.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 4.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 13.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 10.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 22.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 4.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |