Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
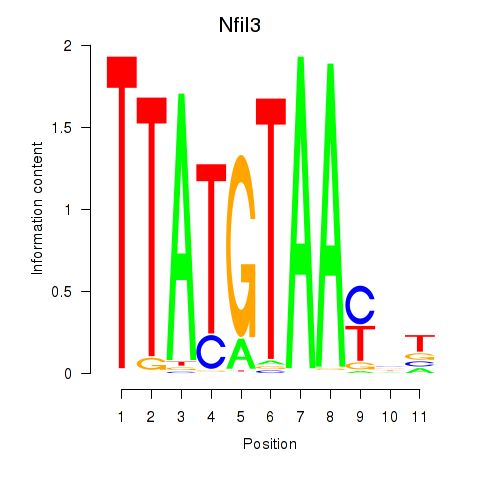
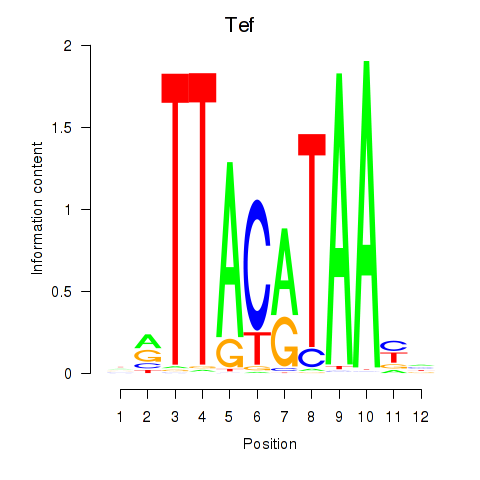
Results for Nfil3_Tef
Z-value: 1.93
Transcription factors associated with Nfil3_Tef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfil3
|
ENSRNOG00000011668 | nuclear factor, interleukin 3 regulated |
|
Tef
|
ENSRNOG00000019383 | TEF, PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tef | rn6_v1_chr7_+_123043503_123043584 | 0.39 | 5.2e-13 | Click! |
| Nfil3 | rn6_v1_chr17_+_12261102_12261102 | 0.28 | 3.8e-07 | Click! |
Activity profile of Nfil3_Tef motif
Sorted Z-values of Nfil3_Tef motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_73529612 | 164.17 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr14_-_19191863 | 121.38 |
ENSRNOT00000003921
|
Alb
|
albumin |
| chr17_+_41798783 | 113.30 |
ENSRNOT00000023519
|
Nrsn1
|
neurensin 1 |
| chr1_+_248428099 | 106.32 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr5_+_120340646 | 105.83 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr4_-_85915099 | 104.03 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr3_+_95715193 | 95.12 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr1_-_256813711 | 90.05 |
ENSRNOT00000021055
|
Rbp4
|
retinol binding protein 4 |
| chr5_-_130085838 | 82.25 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr2_-_41784929 | 82.11 |
ENSRNOT00000086851
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr1_+_224800252 | 80.37 |
ENSRNOT00000024488
|
Slc22a8
|
solute carrier family 22 member 8 |
| chr11_-_81639872 | 78.97 |
ENSRNOT00000047595
ENSRNOT00000090031 ENSRNOT00000081864 |
Hrg
|
histidine-rich glycoprotein |
| chr3_-_44086006 | 77.31 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr15_+_40665041 | 63.26 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr8_-_94563760 | 60.69 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
| chr12_-_29743705 | 57.76 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr11_+_57505005 | 57.22 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr11_+_60072727 | 56.36 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr9_-_82461903 | 54.28 |
ENSRNOT00000026654
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr10_-_90030639 | 52.17 |
ENSRNOT00000090456
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr9_+_73378057 | 48.57 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr17_-_43537293 | 47.75 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr2_-_35104963 | 47.40 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr18_-_26211445 | 46.40 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chrX_-_10218583 | 46.14 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr1_+_229039889 | 45.42 |
ENSRNOT00000054800
|
Glyatl1
|
glycine-N-acyltransferase-like 1 |
| chr1_+_250426158 | 43.30 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr1_+_15642153 | 41.71 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr1_+_84470829 | 41.66 |
ENSRNOT00000025472
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr14_-_3300200 | 40.54 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr11_+_42259761 | 40.52 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr3_+_129018592 | 40.20 |
ENSRNOT00000007274
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr4_-_161850875 | 39.78 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr16_-_45929 | 37.73 |
ENSRNOT00000043153
|
AABR07024439.1
|
|
| chr9_-_8632017 | 37.02 |
ENSRNOT00000043269
|
AABR07066416.1
|
|
| chr14_+_66598259 | 36.85 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr11_+_62584959 | 35.63 |
ENSRNOT00000071065
|
Gramd1c
|
GRAM domain containing 1C |
| chr11_-_81660395 | 35.25 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr1_-_67065797 | 35.14 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr16_+_66380173 | 34.66 |
ENSRNOT00000048862
|
LOC681180
|
similar to Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg) |
| chr9_+_117538346 | 34.61 |
ENSRNOT00000022849
|
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr16_+_66230011 | 34.56 |
ENSRNOT00000074550
|
LOC689479
|
similar to Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg) |
| chr16_+_18690246 | 34.33 |
ENSRNOT00000081484
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr2_+_198321142 | 34.09 |
ENSRNOT00000028760
|
Sv2a
|
synaptic vesicle glycoprotein 2a |
| chr9_+_12475006 | 34.02 |
ENSRNOT00000079703
|
LOC100912293
|
uncharacterized LOC100912293 |
| chrX_+_33884499 | 33.61 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr1_+_217345545 | 33.27 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr16_+_66230841 | 32.91 |
ENSRNOT00000050464
|
LOC689479
|
similar to Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg) |
| chr9_-_9142339 | 32.80 |
ENSRNOT00000046852
|
MGC116197
|
similar to RIKEN cDNA 1700001E04 |
| chr9_+_105943512 | 32.32 |
ENSRNOT00000046409
ENSRNOT00000075578 |
LOC100912293
|
uncharacterized LOC100912293 |
| chr9_-_464390 | 32.23 |
ENSRNOT00000051066
|
LOC302192
|
similar to RIKEN cDNA 1700001E04 |
| chr10_-_90030423 | 32.04 |
ENSRNOT00000092150
|
Mpp2
|
membrane palmitoylated protein 2 |
| chrX_+_9436707 | 31.92 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr1_+_27476375 | 31.71 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr1_+_83163079 | 31.70 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chr20_+_10930518 | 31.36 |
ENSRNOT00000001589
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr1_+_217345154 | 30.62 |
ENSRNOT00000092516
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr4_-_11610518 | 30.54 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr9_-_9143189 | 30.14 |
ENSRNOT00000089904
|
MGC116197
|
similar to RIKEN cDNA 1700001E04 |
| chr4_-_176528110 | 30.07 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr1_+_28454966 | 30.03 |
ENSRNOT00000078841
ENSRNOT00000030327 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr2_+_22000106 | 29.32 |
ENSRNOT00000061812
|
Ankrd34b
|
ankyrin repeat domain 34B |
| chr2_+_252771017 | 28.47 |
ENSRNOT00000075452
ENSRNOT00000055318 |
Ttll7
|
tubulin tyrosine ligase like 7 |
| chr5_-_58163584 | 27.55 |
ENSRNOT00000060594
|
Ccl27
|
C-C motif chemokine ligand 27 |
| chrX_+_22788660 | 27.48 |
ENSRNOT00000071886
|
AABR07037510.1
|
|
| chr11_+_20474483 | 27.20 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr3_+_177310753 | 26.17 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr19_+_15294248 | 26.01 |
ENSRNOT00000024622
|
Ces1f
|
carboxylesterase 1F |
| chr6_+_48452369 | 25.92 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr2_-_216443518 | 25.28 |
ENSRNOT00000022496
|
Amy1a
|
amylase, alpha 1A |
| chr13_-_82753438 | 24.38 |
ENSRNOT00000075948
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr17_+_8489266 | 24.30 |
ENSRNOT00000016252
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_-_138683318 | 23.97 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr18_+_81821127 | 23.89 |
ENSRNOT00000058199
|
Fbxo15
|
F-box protein 15 |
| chr20_-_54517709 | 23.77 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr7_+_110031696 | 23.40 |
ENSRNOT00000012753
|
Khdrbs3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr13_-_111972603 | 23.25 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr9_-_19749145 | 23.12 |
ENSRNOT00000013956
|
Rcan2
|
regulator of calcineurin 2 |
| chr14_-_46153212 | 23.06 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr3_+_124515978 | 22.88 |
ENSRNOT00000028881
|
Prnp
|
prion protein |
| chr1_-_258875572 | 22.65 |
ENSRNOT00000093005
|
Cyp2c13
|
cytochrome P450, family 2, subfamily c, polypeptide 13 |
| chr11_-_81717521 | 22.64 |
ENSRNOT00000058422
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr4_-_68597586 | 22.62 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chr9_+_25410669 | 22.11 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr1_-_67206713 | 22.08 |
ENSRNOT00000048195
|
Vom1r43
|
vomeronasal 1 receptor 43 |
| chr2_-_197991574 | 21.64 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr15_-_47442664 | 21.50 |
ENSRNOT00000072994
|
Prss55
|
protease, serine, 55 |
| chr1_+_147713892 | 21.10 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr4_+_110699557 | 20.63 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr6_-_14523961 | 20.18 |
ENSRNOT00000071402
|
Nrxn1
|
neurexin 1 |
| chr1_-_279277339 | 19.80 |
ENSRNOT00000023667
|
Gfra1
|
GDNF family receptor alpha 1 |
| chr1_-_143535583 | 19.72 |
ENSRNOT00000087785
|
Homer2
|
homer scaffolding protein 2 |
| chr1_-_261669584 | 19.71 |
ENSRNOT00000020568
ENSRNOT00000076555 |
Crtac1
|
cartilage acidic protein 1 |
| chr5_+_58661049 | 19.69 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr1_+_219759183 | 19.33 |
ENSRNOT00000026316
|
Pc
|
pyruvate carboxylase |
| chr18_+_31444472 | 19.19 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr4_+_110700403 | 18.69 |
ENSRNOT00000092379
|
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr12_-_35979193 | 18.48 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr17_-_2278613 | 18.10 |
ENSRNOT00000046525
|
Cntnap3b
|
contactin associated protein-like 3B |
| chr12_+_18679789 | 18.06 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr11_+_88732381 | 17.80 |
ENSRNOT00000078367
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_248649441 | 17.52 |
ENSRNOT00000067165
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr5_+_107369730 | 17.20 |
ENSRNOT00000046837
|
LOC100912314
|
interferon alpha-1-like |
| chr20_-_15334745 | 16.92 |
ENSRNOT00000067633
|
Pcdh15
|
protocadherin 15 |
| chr13_-_57080491 | 16.57 |
ENSRNOT00000017749
ENSRNOT00000086572 ENSRNOT00000060111 |
Cfh
|
complement factor H |
| chr2_-_184263564 | 16.46 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chrX_-_138972684 | 16.30 |
ENSRNOT00000040165
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr1_-_3763392 | 16.23 |
ENSRNOT00000034744
|
Samd5
|
sterile alpha motif domain containing 5 |
| chr5_-_160742479 | 16.00 |
ENSRNOT00000019447
|
Kazn
|
kazrin, periplakin interacting protein |
| chr2_+_247248407 | 15.94 |
ENSRNOT00000082287
|
Unc5c
|
unc-5 netrin receptor C |
| chr10_-_58693754 | 15.25 |
ENSRNOT00000071764
|
Pitpnm3
|
PITPNM family member 3 |
| chr2_+_184230459 | 14.93 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr2_+_3662763 | 14.91 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr7_+_120140460 | 14.76 |
ENSRNOT00000040513
ENSRNOT00000073905 |
Pdxp
|
pyridoxal phosphatase |
| chr2_-_57935334 | 14.47 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr1_-_67175736 | 14.28 |
ENSRNOT00000046788
|
Vom1r44
|
vomeronasal 1 receptor 44 |
| chr2_+_150146234 | 14.16 |
ENSRNOT00000018761
|
Aadac
|
arylacetamide deacetylase |
| chr12_+_18936594 | 13.83 |
ENSRNOT00000081587
|
Spry3
|
sprouty RTK signaling antagonist 3 |
| chr2_-_244335165 | 13.31 |
ENSRNOT00000084860
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr6_-_102047758 | 13.22 |
ENSRNOT00000012101
|
Atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr1_+_17602281 | 13.18 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr18_+_29987206 | 12.69 |
ENSRNOT00000027383
|
Pcdha4
|
protocadherin alpha 4 |
| chr17_-_45154355 | 12.64 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chrX_-_13116743 | 12.34 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr1_-_224698514 | 12.13 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr7_+_73222730 | 12.12 |
ENSRNOT00000007816
|
Erich5
|
glutamate-rich 5 |
| chrX_+_70645270 | 11.63 |
ENSRNOT00000076456
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr3_+_110669312 | 11.54 |
ENSRNOT00000013829
|
Ivd
|
isovaleryl-CoA dehydrogenase |
| chr11_-_1983513 | 11.46 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr1_+_255185629 | 11.29 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr1_+_83103925 | 11.29 |
ENSRNOT00000047540
ENSRNOT00000028196 |
Cyp2b2
|
cytochrome P450, family 2, subfamily b, polypeptide 2 |
| chr15_+_7871497 | 11.24 |
ENSRNOT00000046879
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr1_+_156262031 | 11.23 |
ENSRNOT00000038279
|
Tmem126b
|
transmembrane protein 126B |
| chr14_+_34446616 | 11.21 |
ENSRNOT00000002976
|
Clock
|
clock circadian regulator |
| chr18_+_30010918 | 11.17 |
ENSRNOT00000084132
|
Pcdha4
|
protocadherin alpha 4 |
| chr19_+_17290178 | 10.88 |
ENSRNOT00000060865
|
Aktip
|
AKT interacting protein |
| chr3_-_57957346 | 10.70 |
ENSRNOT00000036728
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr1_+_154579949 | 10.33 |
ENSRNOT00000045048
|
Sytl2
|
synaptotagmin-like 2 |
| chr2_+_251200686 | 10.29 |
ENSRNOT00000019210
|
Col24a1
|
collagen type XXIV alpha 1 chain |
| chr7_-_7189599 | 10.16 |
ENSRNOT00000004458
|
Olr1016
|
olfactory receptor 1016 |
| chr10_-_66873948 | 10.00 |
ENSRNOT00000039261
|
Evi2a
|
ecotropic viral integration site 2A |
| chr2_+_4252496 | 9.24 |
ENSRNOT00000071535
|
RGD1560883
|
similar to KIAA0825 protein |
| chr3_-_146396299 | 9.23 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr4_+_167219728 | 9.05 |
ENSRNOT00000075273
|
Smim10l1
|
small integral membrane protein 10 like 1 |
| chr2_-_53300404 | 8.90 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr20_+_11972381 | 8.89 |
ENSRNOT00000001642
|
Adarb1
|
adenosine deaminase, RNA-specific, B1 |
| chr1_+_61095171 | 8.72 |
ENSRNOT00000042702
|
Vom1r19
|
vomeronasal 1 receptor 19 |
| chrX_+_20216587 | 8.50 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr18_+_30496318 | 8.49 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr3_-_21904133 | 8.39 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr14_+_48740190 | 8.16 |
ENSRNOT00000031638
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_+_61655963 | 7.90 |
ENSRNOT00000040461
|
Coq10b
|
coenzyme Q10B |
| chr3_-_73879647 | 7.53 |
ENSRNOT00000090000
|
Olr510
|
olfactory receptor 510 |
| chr18_-_41389510 | 7.53 |
ENSRNOT00000005476
ENSRNOT00000005446 |
Sema6a
|
semaphorin 6A |
| chr13_-_104080631 | 7.49 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr1_-_141655417 | 7.47 |
ENSRNOT00000068084
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr6_+_73358112 | 7.46 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr10_-_56289882 | 7.42 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr5_+_58995249 | 7.41 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr3_+_79729739 | 7.34 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr16_+_54291251 | 6.89 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr1_+_167944448 | 6.78 |
ENSRNOT00000087667
|
Olr56
|
olfactory receptor 56 |
| chr9_+_16749809 | 6.57 |
ENSRNOT00000028786
|
Cul9
|
cullin 9 |
| chr12_+_8725517 | 6.54 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr2_+_12516702 | 6.53 |
ENSRNOT00000050072
|
Tmem161b
|
transmembrane protein 161B |
| chr10_-_94406949 | 6.51 |
ENSRNOT00000012533
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr20_+_1781320 | 6.43 |
ENSRNOT00000040388
|
Olr1737
|
olfactory receptor 1737 |
| chr13_+_44475970 | 6.41 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chrX_-_77559348 | 6.41 |
ENSRNOT00000047823
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr3_+_160092975 | 6.39 |
ENSRNOT00000080707
|
Pkig
|
cAMP-dependent protein kinase inhibitor gamma |
| chr14_-_72970329 | 6.28 |
ENSRNOT00000006325
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr8_+_82038967 | 6.24 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr1_-_62558033 | 6.19 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr9_-_66483614 | 6.14 |
ENSRNOT00000022047
|
Sumo1
|
small ubiquitin-like modifier 1 |
| chr13_-_95348913 | 6.11 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr7_-_6803318 | 6.10 |
ENSRNOT00000084203
|
Olr954
|
olfactory receptor 954 |
| chr16_-_14300951 | 5.96 |
ENSRNOT00000017038
|
Rgr
|
retinal G protein coupled receptor |
| chr5_-_149987910 | 5.88 |
ENSRNOT00000091637
|
Ptpru
|
protein tyrosine phosphatase, receptor type, U |
| chr16_+_54332660 | 5.86 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr2_-_178297172 | 5.85 |
ENSRNOT00000038543
|
Fnip2
|
folliculin interacting protein 2 |
| chr2_+_46203776 | 5.81 |
ENSRNOT00000074056
|
LOC108350228
|
olfactory receptor 147-like |
| chr14_+_79261092 | 5.80 |
ENSRNOT00000029191
|
LOC680039
|
hypothetical protein LOC680039 |
| chr7_+_130308532 | 5.68 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr1_+_240908483 | 5.62 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr5_-_153184940 | 5.52 |
ENSRNOT00000079536
|
Tmem57
|
transmembrane protein 57 |
| chr4_+_22084954 | 5.51 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr2_+_206314213 | 5.40 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr1_-_43638161 | 5.15 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_70407177 | 5.08 |
ENSRNOT00000049895
|
Os9
|
OS9, endoplasmic reticulum lectin |
| chr3_-_46361092 | 5.06 |
ENSRNOT00000008987
|
Cd302
|
CD302 molecule |
| chr2_-_30634243 | 4.96 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr1_-_282235974 | 4.95 |
ENSRNOT00000054674
|
Sfxn4
|
sideroflexin 4 |
| chr7_-_117364697 | 4.94 |
ENSRNOT00000077314
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr3_-_75089064 | 4.86 |
ENSRNOT00000046167
|
Olr542
|
olfactory receptor 542 |
| chr3_+_93968855 | 4.81 |
ENSRNOT00000014478
|
Fbxo3
|
F-box protein 3 |
| chr6_-_89933168 | 4.78 |
ENSRNOT00000005511
|
AABR07064667.1
|
|
| chr10_-_44471915 | 4.64 |
ENSRNOT00000089901
|
Olr1440
|
olfactory receptor 1440 |
| chr9_+_23596964 | 4.42 |
ENSRNOT00000064279
|
LOC108351994
|
exocrine gland-secreted peptide 1-like |
| chr1_-_53038229 | 4.25 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr1_-_22661377 | 4.21 |
ENSRNOT00000021896
|
Vnn3
|
vanin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfil3_Tef
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.7 | 95.1 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 30.0 | 90.1 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 26.8 | 80.4 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 26.6 | 106.3 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 26.5 | 105.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 26.3 | 79.0 | GO:0097037 | heme export(GO:0097037) |
| 20.2 | 121.4 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) response to platinum ion(GO:0070541) |
| 15.4 | 46.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 10.8 | 43.3 | GO:0016554 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) cytidine to uridine editing(GO:0016554) |
| 8.7 | 60.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 8.1 | 24.4 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 7.8 | 23.3 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 7.6 | 22.9 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 6.9 | 34.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 6.3 | 50.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 6.3 | 25.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 5.8 | 34.6 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 5.5 | 16.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 5.4 | 59.9 | GO:0015747 | urate transport(GO:0015747) |
| 5.4 | 16.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 5.3 | 21.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 4.9 | 63.9 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 4.8 | 19.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 4.6 | 27.6 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 4.5 | 104.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 4.5 | 18.1 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 4.4 | 22.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 4.4 | 39.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 3.8 | 11.5 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 3.6 | 54.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 3.5 | 31.7 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 3.5 | 17.5 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 3.0 | 8.9 | GO:0061744 | motor behavior(GO:0061744) |
| 2.8 | 28.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 2.8 | 19.7 | GO:0099525 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 2.7 | 30.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.7 | 15.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 2.5 | 17.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.5 | 37.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 2.5 | 7.4 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 2.4 | 26.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 2.2 | 11.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 2.1 | 6.4 | GO:0019086 | late viral transcription(GO:0019086) |
| 2.1 | 34.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 2.0 | 41.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 1.9 | 11.3 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 1.9 | 16.9 | GO:0050957 | equilibrioception(GO:0050957) |
| 1.9 | 7.5 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 1.9 | 11.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 1.8 | 23.1 | GO:0007614 | short-term memory(GO:0007614) |
| 1.6 | 63.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 1.6 | 46.9 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 1.6 | 6.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 1.5 | 9.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.5 | 6.1 | GO:0090204 | PML body organization(GO:0030578) protein localization to nuclear pore(GO:0090204) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 1.5 | 13.3 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 1.4 | 17.2 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 1.4 | 14.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 1.4 | 4.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 1.4 | 2.8 | GO:0007172 | signal complex assembly(GO:0007172) |
| 1.4 | 102.0 | GO:0042220 | response to cocaine(GO:0042220) |
| 1.3 | 84.2 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 1.3 | 23.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 1.2 | 16.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 1.2 | 11.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 1.1 | 3.4 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.1 | 42.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 1.1 | 5.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 1.0 | 4.1 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 1.0 | 35.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 1.0 | 3.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.0 | 8.9 | GO:0000255 | allantoin metabolic process(GO:0000255) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 1.0 | 22.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.0 | 21.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.9 | 21.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.9 | 2.7 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.8 | 8.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.8 | 16.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.8 | 10.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.7 | 5.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.7 | 23.4 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.7 | 82.1 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.6 | 7.5 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.6 | 35.2 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.6 | 6.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.6 | 79.9 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.5 | 6.5 | GO:0006983 | ER overload response(GO:0006983) |
| 0.5 | 1.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.5 | 2.6 | GO:2000790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.5 | 7.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 2.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.5 | 3.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.5 | 28.0 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.4 | 31.5 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.4 | 7.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.4 | 10.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.4 | 7.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 1.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.4 | 37.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.4 | 1.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.4 | 1.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 9.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.3 | 4.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 10.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.3 | 72.1 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.3 | 3.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.3 | 5.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 3.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 3.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 12.1 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.3 | 2.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 30.0 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.2 | 2.0 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 4.0 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.2 | 6.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 2.0 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 19.8 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.2 | 7.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.2 | 4.4 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.2 | 2.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.9 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 5.0 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.4 | GO:0006063 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.1 | 1.4 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.1 | 9.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.4 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 98.6 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 0.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 4.3 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.1 | 1.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 2.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 2.9 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 2.9 | GO:0031334 | positive regulation of protein complex assembly(GO:0031334) |
| 0.0 | 4.1 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 20.2 | 60.7 | GO:0098830 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 19.7 | 79.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 12.9 | 77.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 7.2 | 72.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 6.2 | 43.3 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 5.6 | 84.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 5.4 | 48.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 5.0 | 40.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 4.0 | 63.9 | GO:0005883 | neurofilament(GO:0005883) |
| 4.0 | 23.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 3.3 | 16.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 3.1 | 30.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 2.5 | 19.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 2.5 | 31.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 2.2 | 10.9 | GO:0070695 | FHF complex(GO:0070695) |
| 2.2 | 34.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 2.1 | 6.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 2.1 | 57.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 2.0 | 24.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.9 | 3.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.9 | 11.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 1.8 | 8.9 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 1.7 | 50.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.6 | 4.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.5 | 34.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.4 | 19.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.4 | 4.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.1 | 83.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 1.1 | 150.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 1.1 | 14.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.0 | 5.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 1.0 | 2.0 | GO:0043219 | lateral loop(GO:0043219) |
| 1.0 | 5.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 6.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.7 | 260.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.7 | 16.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.6 | 7.5 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.6 | 13.2 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.5 | 16.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.5 | 20.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 18.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 36.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.4 | 30.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 93.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.4 | 3.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.4 | 22.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.4 | 1.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.4 | 19.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.4 | 67.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 77.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.3 | 19.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 18.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 137.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 0.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 11.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 14.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 6.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 26.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 31.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 14.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 1.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 18.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 17.0 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.0 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 4.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 40.5 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.5 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.3 | 106.3 | GO:0005534 | galactose binding(GO:0005534) |
| 11.4 | 34.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 10.0 | 80.4 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 9.5 | 95.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 8.4 | 25.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 8.0 | 39.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 7.8 | 23.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 7.6 | 30.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 7.6 | 45.4 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 6.9 | 90.1 | GO:0019841 | retinol binding(GO:0019841) |
| 6.7 | 60.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 6.6 | 19.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 6.4 | 19.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 6.4 | 121.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 6.0 | 59.9 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 5.8 | 23.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 5.7 | 22.9 | GO:1903135 | cupric ion binding(GO:1903135) |
| 5.7 | 28.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 5.7 | 22.6 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 5.6 | 134.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 4.9 | 14.8 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 4.9 | 63.9 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 4.4 | 17.5 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 3.8 | 30.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 3.7 | 11.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 3.4 | 23.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 3.3 | 16.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 3.3 | 82.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 3.2 | 25.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 3.2 | 15.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 3.2 | 82.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 3.0 | 42.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 2.9 | 35.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 2.9 | 14.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.9 | 20.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 2.8 | 85.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 2.7 | 50.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 2.2 | 8.9 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 2.2 | 8.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 2.2 | 19.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.1 | 10.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 2.0 | 40.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 1.8 | 5.5 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 1.7 | 17.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 1.7 | 34.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 1.6 | 4.9 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 1.3 | 9.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.3 | 7.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.3 | 11.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.2 | 4.8 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 1.2 | 5.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.1 | 16.3 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 1.1 | 44.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.0 | 4.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.0 | 189.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 1.0 | 62.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 1.0 | 3.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.9 | 22.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.9 | 6.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.9 | 18.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.8 | 4.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.8 | 11.5 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.8 | 3.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.8 | 6.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.8 | 76.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.7 | 11.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.7 | 32.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.7 | 36.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.7 | 84.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.6 | 7.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 19.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.6 | 3.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 3.3 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.5 | 3.8 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.5 | 6.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 1.5 | GO:0052723 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.5 | 2.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.4 | 34.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 2.4 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.3 | 2.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.3 | 1.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 15.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 5.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 2.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 8.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 8.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 7.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 8.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.2 | 3.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 0.5 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 0.8 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 3.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 4.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 11.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 3.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 5.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 16.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 5.1 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 5.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.9 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 8.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 22.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 22.0 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 11.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 4.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 3.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 121.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 1.5 | 31.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 1.0 | 95.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.9 | 55.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.8 | 18.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.7 | 110.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.7 | 11.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.6 | 11.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.6 | 16.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.6 | 15.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.6 | 5.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.5 | 6.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.5 | 22.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.4 | 107.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 5.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.4 | 27.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.4 | 6.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 8.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 7.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 3.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 10.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 8.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 6.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.1 | 151.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 9.7 | 106.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 7.3 | 80.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 6.3 | 95.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 4.4 | 105.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 4.2 | 25.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 2.2 | 35.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.2 | 34.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 1.2 | 16.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.1 | 30.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 1.0 | 42.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.9 | 19.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.9 | 33.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.9 | 24.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.8 | 79.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.8 | 30.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.8 | 11.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.8 | 6.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.7 | 6.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.7 | 23.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.6 | 11.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 16.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.5 | 16.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.5 | 13.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 31.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 7.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 18.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.4 | 8.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 10.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 18.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.4 | 6.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.3 | 9.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 17.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 6.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 5.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 4.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 8.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 16.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 8.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 2.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 4.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 4.3 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |