Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
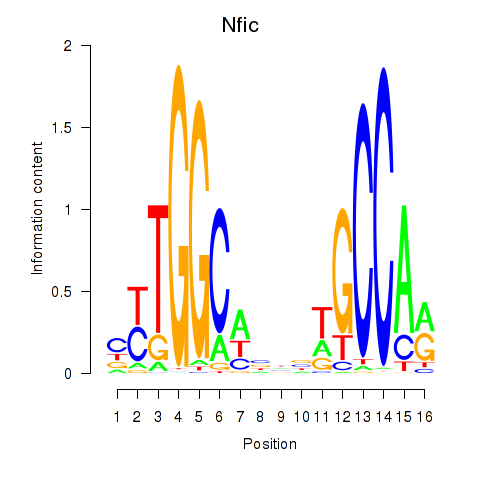
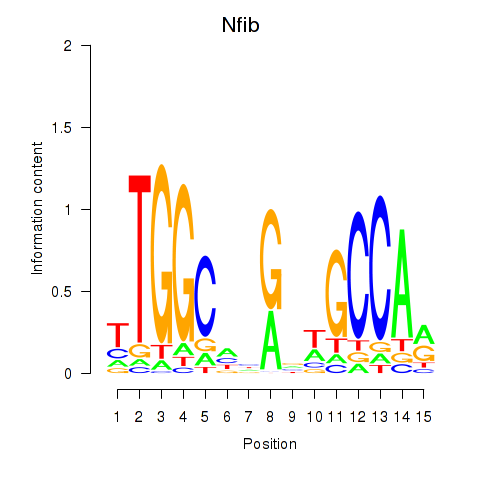
Results for Nfic_Nfib
Z-value: 2.06
Transcription factors associated with Nfic_Nfib
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfic
|
ENSRNOG00000004505 | nuclear factor I/C |
|
Nfib
|
ENSRNOG00000009795 | nuclear factor I/B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfib | rn6_v1_chr5_-_100647298_100647298 | 0.67 | 2.7e-43 | Click! |
| Nfic | rn6_v1_chr7_+_11152038_11152038 | 0.63 | 1.9e-37 | Click! |
Activity profile of Nfic_Nfib motif
Sorted Z-values of Nfic_Nfib motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_27815261 | 182.81 |
ENSRNOT00000032992
|
Klhl33
|
kelch-like family member 33 |
| chr5_-_134526089 | 94.70 |
ENSRNOT00000013321
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr1_-_89543967 | 81.57 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr9_-_82008620 | 71.34 |
ENSRNOT00000023365
|
Prkag3
|
protein kinase AMP-activated non-catalytic subunit gamma 3 |
| chr16_+_54291251 | 66.73 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr10_+_57278307 | 64.19 |
ENSRNOT00000005612
|
Eno3
|
enolase 3 |
| chr5_-_101166651 | 62.48 |
ENSRNOT00000078862
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr1_-_100530183 | 61.21 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr9_-_80167033 | 60.48 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr9_-_20195566 | 59.96 |
ENSRNOT00000015223
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr8_-_130429132 | 59.67 |
ENSRNOT00000026261
|
Hhatl
|
hedgehog acyltransferase-like |
| chr19_-_56677084 | 58.49 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr9_-_80166807 | 57.48 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr4_-_85386231 | 55.73 |
ENSRNOT00000015316
|
Inmt
|
indolethylamine N-methyltransferase |
| chr3_+_11679530 | 54.30 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr5_+_159967839 | 53.61 |
ENSRNOT00000051317
|
Hspb7
|
heat shock protein family B (small) member 7 |
| chr20_+_5008508 | 52.49 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr6_-_129010271 | 52.06 |
ENSRNOT00000075378
|
Serpina10
|
serpin family A member 10 |
| chrX_-_134866210 | 50.54 |
ENSRNOT00000005331
|
Apln
|
apelin |
| chr7_-_144993652 | 50.52 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr7_-_15190796 | 49.40 |
ENSRNOT00000006533
|
Cyp4f1
|
cytochrome P450, family 4, subfamily f, polypeptide 1 |
| chr4_+_44597123 | 47.15 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr1_-_198104109 | 46.31 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr3_+_14467330 | 46.03 |
ENSRNOT00000078939
|
Gsn
|
gelsolin |
| chr4_+_56711049 | 45.73 |
ENSRNOT00000027237
|
Flnc
|
filamin C |
| chr5_+_134492756 | 42.28 |
ENSRNOT00000012888
ENSRNOT00000057095 ENSRNOT00000051385 |
Cyp4a1
|
cytochrome P450, family 4, subfamily a, polypeptide 1 |
| chr1_+_48273611 | 41.77 |
ENSRNOT00000022254
ENSRNOT00000022068 |
Slc22a1
|
solute carrier family 22 member 1 |
| chr8_+_59507990 | 41.72 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chr1_+_80321585 | 40.43 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr12_+_24978483 | 40.03 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr10_+_49231730 | 38.77 |
ENSRNOT00000065335
|
Trim16
|
tripartite motif-containing 16 |
| chr13_-_84331905 | 38.45 |
ENSRNOT00000004965
|
Dusp27
|
dual specificity phosphatase 27 (putative) |
| chr8_-_55177818 | 38.18 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr5_-_93244202 | 37.62 |
ENSRNOT00000075474
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D |
| chr1_-_49844547 | 36.79 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr16_-_74122889 | 35.38 |
ENSRNOT00000025763
|
Plat
|
plasminogen activator, tissue type |
| chr5_-_75005567 | 35.05 |
ENSRNOT00000016068
|
LOC685849
|
hypothetical protein LOC685849 |
| chr19_+_23389375 | 33.15 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr4_+_169147243 | 32.57 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr20_+_6923489 | 32.49 |
ENSRNOT00000093373
ENSRNOT00000000632 |
Pi16
|
peptidase inhibitor 16 |
| chr13_-_80738634 | 32.47 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr8_-_58195884 | 32.38 |
ENSRNOT00000010573
|
Acat1
|
acetyl-CoA acetyltransferase 1 |
| chr9_+_46996156 | 32.21 |
ENSRNOT00000019673
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr4_+_144382945 | 32.17 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr6_+_107245820 | 32.16 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr4_-_100232559 | 32.01 |
ENSRNOT00000016984
|
Vamp5
|
vesicle-associated membrane protein 5 |
| chr1_-_213650247 | 31.34 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr9_+_50526811 | 30.74 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chr4_+_169161585 | 30.63 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr9_+_46997798 | 30.59 |
ENSRNOT00000087112
ENSRNOT00000082408 |
Il1r1
|
interleukin 1 receptor type 1 |
| chr4_+_62220736 | 30.03 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
| chr3_-_110556808 | 29.95 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr11_-_39448503 | 29.78 |
ENSRNOT00000047347
|
LOC102553613
|
SH3 domain-binding glutamic acid-rich protein-like |
| chr14_-_8510138 | 29.21 |
ENSRNOT00000080758
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr18_+_30954609 | 29.08 |
ENSRNOT00000086893
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr7_-_119441487 | 28.73 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr15_+_33632416 | 28.72 |
ENSRNOT00000068212
|
AC130940.1
|
|
| chr10_-_62254287 | 28.68 |
ENSRNOT00000004313
|
Serpinf1
|
serpin family F member 1 |
| chr16_+_54153054 | 28.50 |
ENSRNOT00000090644
ENSRNOT00000014248 |
Fgl1
|
fibrinogen-like 1 |
| chr2_+_68820615 | 27.96 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr16_-_8885797 | 27.90 |
ENSRNOT00000073370
|
RGD1564899
|
similar to chromosome 10 open reading frame 71 |
| chr3_+_109862117 | 27.88 |
ENSRNOT00000083351
ENSRNOT00000070912 |
Thbs1
|
thrombospondin 1 |
| chr3_-_10371240 | 27.31 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr2_+_84645084 | 26.95 |
ENSRNOT00000015448
|
Cmbl
|
carboxymethylenebutenolidase homolog |
| chr10_-_94557764 | 26.83 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chr2_+_95320283 | 26.68 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr2_+_235264219 | 26.66 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr7_+_116632506 | 26.38 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr2_-_235177275 | 26.16 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr2_-_236395067 | 25.97 |
ENSRNOT00000014658
|
Hadh
|
hydroxyacyl-CoA dehydrogenase |
| chr10_+_56662242 | 25.36 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr3_-_175601127 | 25.27 |
ENSRNOT00000081226
|
Lama5
|
laminin subunit alpha 5 |
| chr3_-_160853650 | 25.07 |
ENSRNOT00000018844
|
Matn4
|
matrilin 4 |
| chr2_+_61039053 | 25.03 |
ENSRNOT00000025693
|
Adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr3_+_171213936 | 24.96 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr5_+_164808323 | 24.55 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr1_+_83653234 | 24.29 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr13_+_112031594 | 23.99 |
ENSRNOT00000008440
|
Lamb3
|
laminin subunit beta 3 |
| chr7_+_94375020 | 23.70 |
ENSRNOT00000011904
|
Nov
|
nephroblastoma overexpressed |
| chr4_-_147163467 | 23.53 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr8_+_85355766 | 22.78 |
ENSRNOT00000010583
|
Gcm1
|
glial cells missing homolog 1 |
| chr1_-_167202767 | 22.69 |
ENSRNOT00000055243
|
Art5
|
ADP-ribosyltransferase 5 |
| chr1_+_219764001 | 22.35 |
ENSRNOT00000082388
|
Pc
|
pyruvate carboxylase |
| chr5_+_148257642 | 21.79 |
ENSRNOT00000067663
|
Col16a1
|
collagen type XVI alpha 1 chain |
| chr1_-_241155537 | 21.55 |
ENSRNOT00000034216
ENSRNOT00000073493 |
Mamdc2
|
MAM domain containing 2 |
| chr3_-_8979889 | 21.15 |
ENSRNOT00000065128
|
Crat
|
carnitine O-acetyltransferase |
| chr4_+_44573264 | 21.12 |
ENSRNOT00000080271
|
Cav2
|
caveolin 2 |
| chr1_-_221281180 | 20.53 |
ENSRNOT00000028379
|
Cdc42ep2
|
CDC42 effector protein 2 |
| chr6_+_119519714 | 20.37 |
ENSRNOT00000004955
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr9_-_9985630 | 20.18 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr10_-_98390384 | 20.09 |
ENSRNOT00000065947
|
Abca8a
|
ATP-binding cassette, subfamily A (ABC1), member 8a |
| chr13_+_47454591 | 20.00 |
ENSRNOT00000005791
|
LOC498222
|
similar to specifically androgen-regulated protein |
| chr15_-_52210746 | 19.72 |
ENSRNOT00000046054
|
Bmp1
|
bone morphogenetic protein 1 |
| chr1_+_89008117 | 19.48 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr5_-_61077627 | 19.22 |
ENSRNOT00000015338
|
Shb
|
SH2 domain containing adaptor protein B |
| chr7_+_139698148 | 18.98 |
ENSRNOT00000078579
|
Pfkm
|
phosphofructokinase, muscle |
| chr7_-_15225309 | 18.84 |
ENSRNOT00000075589
ENSRNOT00000066095 |
Cyp4f6
|
cytochrome P450, family 4, subfamily f, polypeptide 6 |
| chr2_-_47281421 | 18.37 |
ENSRNOT00000086114
|
Itga1
|
integrin subunit alpha 1 |
| chr9_+_94702129 | 18.17 |
ENSRNOT00000080930
|
Neu2
|
neuraminidase 2 |
| chr1_-_261986759 | 18.17 |
ENSRNOT00000021257
|
Pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr18_+_32273770 | 18.15 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr9_+_46962288 | 18.14 |
ENSRNOT00000082146
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr1_+_101012822 | 17.76 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chr4_-_115332052 | 17.73 |
ENSRNOT00000017643
|
Clec4f
|
C-type lectin domain family 4, member F |
| chr16_+_59077574 | 17.58 |
ENSRNOT00000090477
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr8_-_22821223 | 17.44 |
ENSRNOT00000014135
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr1_+_144239020 | 16.91 |
ENSRNOT00000032106
|
Adamtsl3
|
ADAMTS-like 3 |
| chr16_+_49266903 | 16.64 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr14_+_80403001 | 16.38 |
ENSRNOT00000012109
|
Cpz
|
carboxypeptidase Z |
| chr13_+_89524329 | 16.18 |
ENSRNOT00000004279
|
Mpz
|
myelin protein zero |
| chr4_+_7158448 | 16.14 |
ENSRNOT00000076953
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr3_+_63510293 | 16.06 |
ENSRNOT00000058093
|
Dfnb59
|
deafness, autosomal recessive 59 |
| chr16_-_40025401 | 15.91 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr8_+_95968652 | 15.68 |
ENSRNOT00000015057
|
Nt5e
|
5' nucleotidase, ecto |
| chr5_+_140888973 | 15.66 |
ENSRNOT00000020524
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr1_-_215033460 | 15.65 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr9_-_100306194 | 15.51 |
ENSRNOT00000087584
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr10_-_88036040 | 15.49 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr10_-_104659188 | 15.22 |
ENSRNOT00000010887
|
Trim47
|
tripartite motif-containing 47 |
| chr7_-_139907640 | 15.21 |
ENSRNOT00000045473
|
Zfp641
|
zinc finger protein 641 |
| chr1_-_170318935 | 15.19 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr7_-_31824064 | 15.15 |
ENSRNOT00000011494
ENSRNOT00000080824 |
Slc25a3
|
solute carrier family 25 member 3 |
| chr10_+_86950557 | 15.10 |
ENSRNOT00000014153
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr12_-_51250230 | 15.05 |
ENSRNOT00000029508
|
Mn1
|
meningioma 1 |
| chr8_-_22821397 | 15.01 |
ENSRNOT00000045488
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr9_+_16924520 | 14.86 |
ENSRNOT00000025094
|
Slc22a7
|
solute carrier family 22 member 7 |
| chr4_+_62019970 | 14.85 |
ENSRNOT00000013133
|
LOC100910708
|
aldose reductase-related protein 1-like |
| chr1_-_222350173 | 14.77 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr17_+_80882666 | 14.73 |
ENSRNOT00000024430
|
Vim
|
vimentin |
| chr12_-_46809849 | 14.55 |
ENSRNOT00000081134
|
Pxn
|
paxillin |
| chr1_-_175796040 | 14.45 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chr6_-_109935533 | 14.43 |
ENSRNOT00000013516
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr3_+_112428395 | 14.42 |
ENSRNOT00000079109
ENSRNOT00000048141 |
Stard9
|
StAR-related lipid transfer domain containing 9 |
| chr2_-_147392062 | 14.36 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr9_+_82596355 | 14.34 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr2_-_195888216 | 14.33 |
ENSRNOT00000056436
|
Tuft1
|
tuftelin 1 |
| chrX_+_68771100 | 14.06 |
ENSRNOT00000043872
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr3_+_160207913 | 13.93 |
ENSRNOT00000014346
|
Wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr8_-_62458301 | 13.92 |
ENSRNOT00000021653
|
Cyp1a2
|
cytochrome P450, family 1, subfamily a, polypeptide 2 |
| chr1_-_126211439 | 13.65 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr5_-_171710312 | 13.38 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chr18_+_32336102 | 13.38 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr11_-_69223158 | 13.20 |
ENSRNOT00000067060
|
Mylk
|
myosin light chain kinase |
| chr1_+_198199622 | 13.14 |
ENSRNOT00000026688
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr19_+_61332351 | 13.11 |
ENSRNOT00000014492
|
Nrp1
|
neuropilin 1 |
| chr1_+_91433030 | 13.08 |
ENSRNOT00000015205
|
Slc7a10
|
solute carrier family 7 member 10 |
| chr1_+_221099998 | 13.04 |
ENSRNOT00000028262
|
Ltbp3
|
latent transforming growth factor beta binding protein 3 |
| chr6_-_62798384 | 12.88 |
ENSRNOT00000093200
|
Ndufa10l1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10-like 1 |
| chr8_-_115140080 | 12.83 |
ENSRNOT00000015851
|
Acy1
|
aminoacylase 1 |
| chr1_+_148240504 | 12.78 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr13_-_52088780 | 12.74 |
ENSRNOT00000008754
|
Elf3
|
E74-like factor 3 |
| chr5_-_4975436 | 12.47 |
ENSRNOT00000062006
|
Xkr9
|
XK related 9 |
| chr6_-_127534247 | 12.32 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr10_-_104575890 | 12.28 |
ENSRNOT00000050223
|
H3f3b
|
H3 histone family member 3B |
| chr9_-_55256340 | 12.13 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr4_-_145300177 | 12.12 |
ENSRNOT00000029728
|
Camk1
|
calcium/calmodulin-dependent protein kinase I |
| chr4_+_145427367 | 12.04 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr7_+_142776580 | 11.62 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr13_+_26970896 | 11.59 |
ENSRNOT00000003501
|
Serpinb12
|
serpin family B member 12 |
| chr17_+_72209373 | 11.58 |
ENSRNOT00000064802
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr8_+_130401470 | 11.55 |
ENSRNOT00000043346
|
Zbtb47
|
zinc finger and BTB domain containing 47 |
| chr1_-_222177421 | 11.49 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr1_-_194769524 | 11.36 |
ENSRNOT00000025988
|
Nupr1
|
nuclear protein 1, transcriptional regulator |
| chr1_+_171797516 | 11.34 |
ENSRNOT00000088110
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr1_+_214778496 | 11.10 |
ENSRNOT00000028967
|
Muc5b
|
mucin 5B, oligomeric mucus/gel-forming |
| chr7_-_117151256 | 11.00 |
ENSRNOT00000078846
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr9_+_51302151 | 10.94 |
ENSRNOT00000085908
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr13_-_90832469 | 10.87 |
ENSRNOT00000086508
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr5_+_113725717 | 10.76 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr1_-_188713270 | 10.71 |
ENSRNOT00000082192
ENSRNOT00000065892 |
Gprc5b
|
G protein-coupled receptor, class C, group 5, member B |
| chr7_-_117151694 | 10.71 |
ENSRNOT00000051294
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr8_-_21995806 | 10.60 |
ENSRNOT00000028034
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr2_+_233615739 | 10.51 |
ENSRNOT00000051009
|
Pitx2
|
paired-like homeodomain 2 |
| chr11_+_30363280 | 10.48 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr7_-_117679219 | 10.47 |
ENSRNOT00000071522
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr1_-_162385575 | 10.45 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr1_-_12952906 | 10.45 |
ENSRNOT00000078193
|
LOC100360362
|
hypothetical protein LOC100360362 |
| chr3_+_98297554 | 10.41 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr5_+_76812931 | 10.40 |
ENSRNOT00000059458
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr5_-_33182147 | 10.31 |
ENSRNOT00000080358
|
Maged2
|
MAGE family member D2 |
| chr13_+_26970660 | 10.30 |
ENSRNOT00000083145
|
Serpinb12
|
serpin family B member 12 |
| chr2_+_143951838 | 10.22 |
ENSRNOT00000079544
|
LOC103691556
|
mothers against decapentaplegic homolog 9 |
| chr13_-_80775230 | 10.21 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr5_-_126096068 | 10.19 |
ENSRNOT00000036793
|
AABR07073134.1
|
|
| chr2_+_251863069 | 10.18 |
ENSRNOT00000036282
|
Syde2
|
synapse defective Rho GTPase homolog 2 |
| chrX_+_143097525 | 10.09 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr7_-_143420027 | 9.89 |
ENSRNOT00000082863
|
Krt2
|
keratin 2 |
| chr3_+_5624506 | 9.76 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr8_-_105462141 | 9.69 |
ENSRNOT00000066731
ENSRNOT00000078760 |
Clstn2
|
calsyntenin 2 |
| chr7_+_2752680 | 9.62 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr5_+_58144705 | 9.61 |
ENSRNOT00000019886
|
Galt
|
galactose-1-phosphate uridylyltransferase |
| chr15_-_46367302 | 9.61 |
ENSRNOT00000032089
|
Fdft1
|
farnesyl diphosphate farnesyl transferase 1 |
| chr16_-_7408265 | 9.51 |
ENSRNOT00000035009
|
Dnah1
|
dynein, axonemal, heavy chain 1 |
| chr3_-_8766433 | 9.39 |
ENSRNOT00000021865
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr3_-_48535909 | 9.34 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr4_-_60358562 | 9.31 |
ENSRNOT00000018001
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr13_-_68360664 | 9.29 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr3_-_161067358 | 9.29 |
ENSRNOT00000055160
|
Wfdc8
|
WAP four-disulfide core domain 8 |
| chr11_+_31002451 | 9.29 |
ENSRNOT00000002838
|
Eva1c
|
eva-1 homolog C |
| chr9_+_81615251 | 9.25 |
ENSRNOT00000081106
ENSRNOT00000020049 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfic_Nfib
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.5 | 118.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 27.2 | 81.6 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 27.0 | 80.9 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 22.8 | 68.3 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 18.1 | 54.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 15.3 | 46.0 | GO:1903921 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 14.6 | 58.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 13.3 | 40.0 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 13.0 | 13.0 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 12.9 | 90.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 12.6 | 50.5 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 11.3 | 56.6 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 11.3 | 33.9 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 11.0 | 33.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 10.8 | 32.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 10.8 | 32.4 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) ketone body catabolic process(GO:0046952) metanephric proximal tubule development(GO:0072237) |
| 10.7 | 32.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 10.6 | 42.3 | GO:0048252 | lauric acid metabolic process(GO:0048252) |
| 9.9 | 59.7 | GO:1903059 | regulation of protein lipidation(GO:1903059) |
| 9.8 | 88.2 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 9.5 | 38.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 9.3 | 27.9 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) regulation of cGMP-mediated signaling(GO:0010752) |
| 9.1 | 27.3 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 8.9 | 26.7 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 8.8 | 35.4 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 8.8 | 26.4 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 8.2 | 24.5 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 7.9 | 23.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) negative regulation of sensory perception of pain(GO:1904057) |
| 7.1 | 28.6 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 6.8 | 61.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 6.6 | 46.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 6.6 | 19.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 6.5 | 38.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 5.8 | 17.4 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 5.4 | 32.5 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 5.3 | 21.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 5.2 | 15.7 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 4.8 | 14.4 | GO:1905071 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 4.6 | 22.8 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 4.5 | 18.2 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 4.4 | 13.1 | GO:0038189 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 4.3 | 42.7 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 4.2 | 41.8 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 4.2 | 16.6 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 4.0 | 24.3 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 4.0 | 28.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 4.0 | 55.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 3.8 | 19.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 3.8 | 26.8 | GO:0015871 | choline transport(GO:0015871) |
| 3.7 | 14.7 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 3.6 | 10.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 3.5 | 10.5 | GO:0021539 | subthalamus development(GO:0021539) deltoid tuberosity development(GO:0035993) pulmonary vein morphogenesis(GO:0060577) |
| 3.5 | 10.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 3.5 | 13.9 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 3.4 | 23.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 3.4 | 20.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 3.3 | 62.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 3.3 | 91.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 3.2 | 9.6 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 3.2 | 28.7 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) cellular response to cobalt ion(GO:0071279) |
| 3.2 | 31.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 3.1 | 30.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 2.8 | 8.3 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 2.7 | 21.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 2.6 | 31.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 2.5 | 15.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 2.5 | 7.4 | GO:0006214 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 2.5 | 12.3 | GO:0060623 | pericentric heterochromatin assembly(GO:0031508) regulation of chromosome condensation(GO:0060623) |
| 2.2 | 17.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 2.1 | 10.5 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 2.1 | 43.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 2.0 | 8.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 2.0 | 12.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 2.0 | 7.9 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 1.9 | 13.4 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 1.9 | 13.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 1.9 | 9.4 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 1.9 | 5.6 | GO:0090648 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.9 | 13.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 1.8 | 9.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 1.8 | 47.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 1.8 | 7.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 1.8 | 10.6 | GO:1903142 | negative regulation of excitatory postsynaptic potential(GO:0090394) positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.8 | 17.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 1.7 | 67.8 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 1.7 | 6.9 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 1.7 | 20.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 1.7 | 13.7 | GO:0071000 | response to magnetism(GO:0071000) |
| 1.7 | 23.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 1.7 | 8.4 | GO:0060282 | negative regulation of cardiac muscle contraction(GO:0055118) positive regulation of oocyte development(GO:0060282) |
| 1.7 | 6.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.6 | 8.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 1.6 | 44.1 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 1.6 | 4.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 1.5 | 15.5 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 1.5 | 10.7 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 1.5 | 15.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 1.5 | 4.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.4 | 14.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 1.4 | 4.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.4 | 8.6 | GO:0061484 | hematopoietic stem cell migration(GO:0035701) hematopoietic stem cell homeostasis(GO:0061484) |
| 1.4 | 71.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 1.4 | 9.8 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 1.4 | 9.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.3 | 3.9 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 1.3 | 10.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.2 | 3.7 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 1.2 | 3.7 | GO:0097187 | dentinogenesis(GO:0097187) |
| 1.2 | 37.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.2 | 4.8 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 1.2 | 3.6 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 1.2 | 7.0 | GO:1902617 | response to fluoride(GO:1902617) |
| 1.1 | 28.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 1.1 | 2.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 1.0 | 30.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 1.0 | 12.0 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 1.0 | 9.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 1.0 | 25.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 1.0 | 14.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) |
| 1.0 | 47.8 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.9 | 8.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.9 | 10.8 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.9 | 9.9 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.9 | 11.4 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.9 | 7.8 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.9 | 4.3 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.8 | 14.4 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.8 | 45.7 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.8 | 3.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.8 | 22.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.8 | 16.2 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.8 | 20.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.8 | 5.5 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.8 | 1.6 | GO:0060729 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) intestinal epithelial structure maintenance(GO:0060729) |
| 0.8 | 17.1 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.8 | 7.7 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.8 | 8.5 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.8 | 5.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.7 | 34.7 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.7 | 19.7 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.7 | 2.9 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.7 | 9.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.7 | 35.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.7 | 24.0 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.7 | 2.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.7 | 15.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.6 | 23.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.6 | 5.0 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.6 | 12.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.6 | 2.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.6 | 8.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.6 | 5.3 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.6 | 12.0 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.5 | 1.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 3.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.5 | 6.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) intestinal epithelial cell development(GO:0060576) |
| 0.5 | 1.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.5 | 5.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.5 | 2.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 17.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.5 | 5.5 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.5 | 12.3 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.4 | 1.8 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.4 | 1.8 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.4 | 2.6 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) response to anoxia(GO:0034059) |
| 0.4 | 26.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 11.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.4 | 9.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.4 | 6.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 4.1 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.4 | 1.9 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.4 | 11.5 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.4 | 10.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.4 | 3.5 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.3 | 39.5 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.3 | 4.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.3 | 10.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.3 | 13.5 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.3 | 25.2 | GO:0009408 | response to heat(GO:0009408) |
| 0.3 | 3.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.3 | 1.5 | GO:1903431 | positive regulation of neuron maturation(GO:0014042) positive regulation of cell maturation(GO:1903431) |
| 0.3 | 4.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 1.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.3 | 2.7 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.3 | 1.8 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 4.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.3 | 40.3 | GO:0071559 | response to transforming growth factor beta(GO:0071559) |
| 0.2 | 17.5 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.2 | 3.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 9.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.2 | 1.8 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.2 | 7.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 0.9 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 0.4 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.2 | 4.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 1.8 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 8.2 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.2 | 2.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 3.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.2 | 7.5 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.2 | 6.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 2.9 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.2 | 1.9 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 6.1 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 | 3.4 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 3.7 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 20.2 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.1 | 18.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.8 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.9 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 5.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 3.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 1.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 2.4 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 1.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 4.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 6.9 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.4 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 25.7 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.1 | 0.2 | GO:0071348 | response to interleukin-11(GO:0071105) cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.3 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.1 | 5.1 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.0 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 8.0 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 2.8 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 6.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 3.2 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 30.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0001842 | neural fold formation(GO:0001842) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.7 | 118.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 12.7 | 76.1 | GO:0030478 | actin cap(GO:0030478) |
| 10.7 | 64.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 8.0 | 40.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 6.3 | 19.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 6.3 | 25.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 5.2 | 47.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 5.1 | 61.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 4.0 | 27.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 4.0 | 71.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 3.6 | 28.7 | GO:0043203 | axon hillock(GO:0043203) |
| 3.0 | 24.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 3.0 | 8.9 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 2.9 | 14.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 2.5 | 17.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 2.4 | 9.5 | GO:0036156 | inner dynein arm(GO:0036156) |
| 2.4 | 77.8 | GO:0008305 | integrin complex(GO:0008305) |
| 2.3 | 6.9 | GO:1990032 | parallel fiber(GO:1990032) |
| 2.2 | 6.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 2.1 | 53.0 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 2.0 | 31.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.8 | 27.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.7 | 32.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.7 | 5.0 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 1.6 | 13.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 1.5 | 12.3 | GO:0001740 | Barr body(GO:0001740) |
| 1.5 | 6.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.5 | 33.1 | GO:0010369 | chromocenter(GO:0010369) |
| 1.5 | 26.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.4 | 11.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 1.3 | 64.5 | GO:0016235 | aggresome(GO:0016235) |
| 1.3 | 9.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.2 | 45.7 | GO:0043034 | costamere(GO:0043034) |
| 1.2 | 21.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 13.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.1 | 3.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.1 | 101.7 | GO:0005604 | basement membrane(GO:0005604) |
| 1.0 | 8.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.8 | 7.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.8 | 21.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.7 | 7.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.7 | 2.9 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.7 | 52.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.7 | 159.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.6 | 9.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.6 | 6.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.6 | 8.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.6 | 32.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.5 | 13.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.4 | 31.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 78.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.4 | 46.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 10.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 3.5 | GO:0032437 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.4 | 14.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 118.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.4 | 272.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.3 | 7.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 4.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 3.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 1.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.3 | 1.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.3 | 319.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.3 | 10.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 15.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 11.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 25.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 1.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 8.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 17.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 1.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 9.1 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 7.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 39.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 5.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 10.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 9.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.2 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 8.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 17.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 2.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 31.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 5.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 27.0 | 80.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 16.3 | 81.6 | GO:0005534 | galactose binding(GO:0005534) |
| 16.3 | 49.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 15.4 | 46.3 | GO:0050656 | steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 14.8 | 133.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 13.9 | 41.8 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 11.5 | 46.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 10.8 | 32.4 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 10.7 | 64.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 10.6 | 42.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 9.4 | 28.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 8.9 | 26.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 7.6 | 22.7 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 7.2 | 79.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 7.0 | 55.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 6.8 | 81.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 6.1 | 61.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 6.1 | 116.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 5.8 | 40.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.6 | 27.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 5.5 | 49.4 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 4.9 | 19.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 4.8 | 71.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 4.7 | 19.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 4.5 | 18.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 4.4 | 26.4 | GO:0035473 | lipase binding(GO:0035473) |
| 4.3 | 12.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 4.3 | 38.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 3.7 | 18.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 3.6 | 42.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 3.4 | 20.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 3.2 | 69.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 3.1 | 9.4 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 2.9 | 14.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 2.6 | 21.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 2.6 | 26.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 2.5 | 7.4 | GO:0017113 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 2.4 | 7.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 2.2 | 44.4 | GO:0031005 | filamin binding(GO:0031005) |
| 2.2 | 8.8 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 2.1 | 25.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 2.1 | 10.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 2.1 | 66.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 2.1 | 41.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 2.0 | 9.9 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 2.0 | 33.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 2.0 | 13.7 | GO:0071253 | connexin binding(GO:0071253) |
| 1.9 | 11.6 | GO:0098821 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 1.9 | 9.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.8 | 9.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 1.8 | 16.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 1.8 | 10.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.7 | 17.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.7 | 12.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.7 | 6.9 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.7 | 6.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.7 | 9.9 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.6 | 28.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.6 | 13.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 1.6 | 8.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.6 | 4.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.6 | 64.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 1.6 | 32.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 1.5 | 10.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.5 | 21.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 1.5 | 8.9 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 1.4 | 11.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.4 | 37.1 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.3 | 5.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 1.3 | 49.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.2 | 25.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 1.2 | 6.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.2 | 40.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 1.2 | 135.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 1.1 | 4.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.1 | 15.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.1 | 3.3 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.1 | 13.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.1 | 13.9 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 1.1 | 10.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 1.1 | 10.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 1.0 | 31.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.0 | 45.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 1.0 | 13.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.0 | 14.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.9 | 33.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.9 | 3.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 24.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.8 | 13.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.8 | 10.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.8 | 5.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.8 | 3.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.8 | 32.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.6 | 49.0 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.6 | 15.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.6 | 3.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.6 | 12.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.6 | 11.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.6 | 1.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.6 | 16.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.6 | 13.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.6 | 8.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.6 | 37.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.6 | 7.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 56.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.6 | 3.9 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.5 | 8.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.5 | 8.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.5 | 30.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.5 | 7.9 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.5 | 3.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 75.9 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.4 | 7.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 2.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 12.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.4 | 1.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 12.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 15.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 1.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 3.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.3 | 1.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 1.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.3 | 0.9 | GO:0008396 | oxysterol 7-alpha-hydroxylase activity(GO:0008396) |
| 0.2 | 2.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 15.0 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.2 | 2.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 4.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 5.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 20.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 11.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 11.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.2 | 8.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 2.6 | GO:0017128 | intracellular calcium activated chloride channel activity(GO:0005229) phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 8.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 8.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 28.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.5 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 1.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 4.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 3.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 15.9 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 5.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 6.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 1.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 7.4 | GO:0042626 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 1.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 5.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 3.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 3.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 30.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 8.1 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 98.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 4.0 | 40.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 3.6 | 68.9 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 2.9 | 49.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 2.6 | 80.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 2.4 | 89.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 2.0 | 76.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 2.0 | 399.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 1.9 | 93.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 1.7 | 26.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 1.5 | 35.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.5 | 13.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 1.4 | 24.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 1.0 | 15.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 1.0 | 50.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.9 | 10.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.9 | 36.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 13.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.7 | 173.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.7 | 31.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.7 | 12.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.6 | 13.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.6 | 21.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.5 | 28.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.5 | 10.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.4 | 61.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 10.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.3 | 6.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 5.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 10.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 7.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 9.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 11.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 1.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 7.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 7.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 4.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 71.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 7.4 | 66.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 6.3 | 133.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 4.6 | 13.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 3.6 | 46.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 3.2 | 29.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 3.1 | 52.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 2.7 | 37.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 2.6 | 76.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 2.6 | 47.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 2.4 | 31.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 2.3 | 135.4 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 2.2 | 42.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 2.1 | 83.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.9 | 83.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.9 | 14.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.9 | 26.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.8 | 32.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.7 | 30.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.7 | 80.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 1.5 | 28.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 1.3 | 105.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 1.3 | 10.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 1.2 | 19.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 1.1 | 13.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.0 | 21.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 1.0 | 22.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.9 | 16.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.9 | 10.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.7 | 7.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.7 | 11.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.7 | 11.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.7 | 22.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 26.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.6 | 8.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.6 | 22.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.6 | 26.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.6 | 6.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.5 | 1.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.5 | 4.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 6.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 42.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.5 | 21.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.5 | 21.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 13.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 8.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 24.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 9.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 5.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 52.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 5.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 6.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 5.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 5.4 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.2 | 3.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 8.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 3.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 3.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.1 | 1.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 10.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 4.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.8 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |