Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
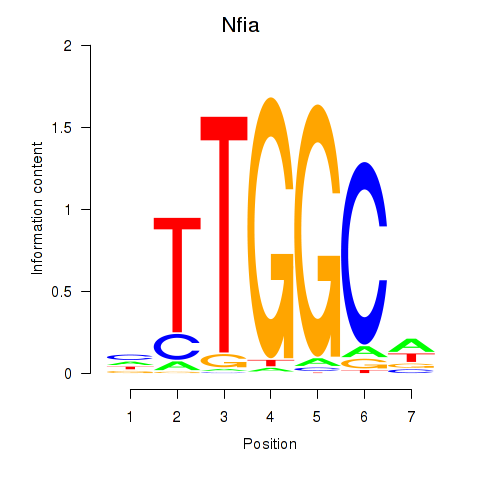
Results for Nfia
Z-value: 3.04
Transcription factors associated with Nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfia
|
ENSRNOG00000006966 | nuclear factor I/A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfia | rn6_v1_chr5_+_116420690_116420690 | 0.48 | 1.4e-19 | Click! |
Activity profile of Nfia motif
Sorted Z-values of Nfia motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_159775643 | 147.09 |
ENSRNOT00000010939
|
Jph2
|
junctophilin 2 |
| chr1_-_213650247 | 119.80 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr15_+_4077951 | 115.78 |
ENSRNOT00000085266
|
Myoz1
|
myozenin 1 |
| chr15_-_27815261 | 105.62 |
ENSRNOT00000032992
|
Klhl33
|
kelch-like family member 33 |
| chr10_+_53778662 | 99.40 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr10_+_86337728 | 98.90 |
ENSRNOT00000085408
|
Tcap
|
titin-cap |
| chr3_-_63568464 | 97.24 |
ENSRNOT00000068494
|
AABR07052585.2
|
|
| chr9_-_73958480 | 97.19 |
ENSRNOT00000017838
|
Myl1
|
myosin, light chain 1 |
| chr17_+_47397558 | 93.59 |
ENSRNOT00000085923
|
Epdr1
|
ependymin related 1 |
| chrX_-_64715823 | 88.22 |
ENSRNOT00000076297
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr10_+_53818818 | 83.17 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr1_-_199624783 | 81.69 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr1_-_104157855 | 81.42 |
ENSRNOT00000019311
|
Csrp3
|
cysteine and glycine rich protein 3 |
| chr7_-_118108864 | 81.00 |
ENSRNOT00000006184
|
Mb
|
myoglobin |
| chr13_-_82005741 | 78.54 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr5_+_140888973 | 78.16 |
ENSRNOT00000020524
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr1_-_104166367 | 77.78 |
ENSRNOT00000092211
|
Csrp3
|
cysteine and glycine rich protein 3 |
| chr13_-_90602365 | 76.19 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr2_-_233743866 | 75.37 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chrX_+_53053609 | 74.96 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr8_+_55178289 | 72.53 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr13_-_82006005 | 71.89 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr10_+_54352270 | 67.78 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr10_+_53740841 | 66.84 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr17_-_389967 | 66.41 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr3_-_105512939 | 66.03 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr3_-_37803112 | 62.33 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr18_+_35574002 | 61.97 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr14_-_86047162 | 61.14 |
ENSRNOT00000018227
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr5_+_152533349 | 60.36 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chrX_+_124631881 | 60.34 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr13_-_52197205 | 58.66 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr7_+_38819771 | 58.12 |
ENSRNOT00000006109
|
Lum
|
lumican |
| chr14_-_62595854 | 56.03 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr1_-_100530183 | 55.77 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr2_-_45518502 | 54.14 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr3_-_8979889 | 53.87 |
ENSRNOT00000065128
|
Crat
|
carnitine O-acetyltransferase |
| chr1_-_49844547 | 52.56 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr5_+_164808323 | 51.48 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr13_+_87986240 | 51.38 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr6_-_127248372 | 50.96 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr2_-_184993341 | 47.90 |
ENSRNOT00000071580
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr12_-_6740714 | 47.51 |
ENSRNOT00000001205
|
Medag
|
mesenteric estrogen-dependent adipogenesis |
| chr4_-_147163467 | 46.77 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr2_+_104416972 | 46.76 |
ENSRNOT00000017125
|
Trim55
|
tripartite motif-containing 55 |
| chr9_-_82008620 | 46.58 |
ENSRNOT00000023365
|
Prkag3
|
protein kinase AMP-activated non-catalytic subunit gamma 3 |
| chrX_-_142248369 | 46.42 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr2_+_204512302 | 46.05 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr10_+_39666991 | 45.71 |
ENSRNOT00000030760
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr9_-_88356716 | 44.80 |
ENSRNOT00000077503
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr4_+_7158970 | 44.68 |
ENSRNOT00000037446
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr1_+_261281543 | 43.79 |
ENSRNOT00000018890
|
Ankrd2
|
ankyrin repeat domain 2 |
| chr5_-_160179978 | 42.81 |
ENSRNOT00000022820
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr7_+_94375020 | 42.70 |
ENSRNOT00000011904
|
Nov
|
nephroblastoma overexpressed |
| chr6_+_98284170 | 42.19 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr5_-_164714145 | 41.97 |
ENSRNOT00000055680
|
LOC100911485
|
mitofusin-2-like |
| chr9_-_88357182 | 41.81 |
ENSRNOT00000041176
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr4_+_155321553 | 41.59 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr2_-_184951950 | 40.27 |
ENSRNOT00000093486
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr4_+_7076759 | 40.05 |
ENSRNOT00000066598
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr10_-_109729019 | 39.18 |
ENSRNOT00000054959
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chrX_-_116792707 | 39.15 |
ENSRNOT00000049482
|
Amot
|
angiomotin |
| chrX_+_131617798 | 38.90 |
ENSRNOT00000074384
|
Prr32
|
proline rich 32 |
| chr17_-_65955606 | 38.62 |
ENSRNOT00000067949
ENSRNOT00000023601 |
Ryr2
|
ryanodine receptor 2 |
| chr1_-_226935689 | 37.96 |
ENSRNOT00000038807
|
Tmem109
|
transmembrane protein 109 |
| chr1_-_215536980 | 37.26 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr5_+_115649046 | 37.09 |
ENSRNOT00000041328
|
LOC108351137
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr10_+_16970626 | 37.06 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr16_+_2537248 | 36.64 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr4_+_7158448 | 36.59 |
ENSRNOT00000076953
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr18_-_63825408 | 36.51 |
ENSRNOT00000043709
|
RGD1562758
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr4_+_180062799 | 36.28 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr13_+_113691932 | 36.04 |
ENSRNOT00000084709
|
Cd34
|
CD34 molecule |
| chr14_+_6994190 | 35.85 |
ENSRNOT00000020357
|
Sparcl1
|
SPARC like 1 |
| chr17_-_84614228 | 35.35 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chr8_-_22542997 | 35.32 |
ENSRNOT00000061009
|
Tmed1
|
transmembrane p24 trafficking protein 1 |
| chr8_-_116349896 | 35.04 |
ENSRNOT00000087306
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr20_-_2701637 | 34.90 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr17_-_14717420 | 34.80 |
ENSRNOT00000071131
|
LOC100910978
|
extracellular matrix protein 2-like |
| chrX_+_77278240 | 34.44 |
ENSRNOT00000085653
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr17_+_15966234 | 34.28 |
ENSRNOT00000084304
|
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr8_+_130009573 | 33.67 |
ENSRNOT00000026106
|
Trak1
|
trafficking kinesin protein 1 |
| chr13_+_83073544 | 33.59 |
ENSRNOT00000066119
ENSRNOT00000079796 ENSRNOT00000077070 |
Dpt
|
dermatopontin |
| chr1_-_82452281 | 33.39 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr16_+_25773602 | 33.33 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr6_-_8344897 | 33.06 |
ENSRNOT00000082353
|
Prepl
|
prolyl endopeptidase-like |
| chrX_-_116792864 | 32.96 |
ENSRNOT00000090918
|
Amot
|
angiomotin |
| chr11_+_70687500 | 32.64 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr10_+_13836128 | 32.27 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr8_+_117268337 | 32.17 |
ENSRNOT00000072098
ENSRNOT00000083578 |
Lamb2
|
laminin subunit beta 2 |
| chr4_-_97784842 | 32.15 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr6_-_8344574 | 32.01 |
ENSRNOT00000009660
|
Prepl
|
prolyl endopeptidase-like |
| chr10_+_89251370 | 31.88 |
ENSRNOT00000076820
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr3_+_57770948 | 31.40 |
ENSRNOT00000038429
|
AC107446.2
|
|
| chr8_-_95387363 | 31.32 |
ENSRNOT00000014657
|
Tbx18
|
T-box18 |
| chr20_+_5008508 | 31.20 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr5_-_152458023 | 31.00 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr10_+_65767930 | 30.70 |
ENSRNOT00000039954
|
Vtn
|
vitronectin |
| chr3_+_148386189 | 30.39 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr12_-_23954431 | 30.35 |
ENSRNOT00000001958
|
Mdh2
|
malate dehydrogenase 2 |
| chr15_-_52214616 | 30.22 |
ENSRNOT00000015035
|
Sftpc
|
surfactant protein C |
| chr13_+_83073866 | 29.97 |
ENSRNOT00000075996
|
Dpt
|
dermatopontin |
| chr2_+_188528979 | 29.94 |
ENSRNOT00000087934
|
Thbs3
|
thrombospondin 3 |
| chr1_-_259484569 | 29.83 |
ENSRNOT00000021289
ENSRNOT00000021114 ENSRNOT00000021229 ENSRNOT00000021347 ENSRNOT00000021226 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr15_-_108120279 | 29.74 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr4_+_145427367 | 29.62 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr5_+_104698040 | 29.57 |
ENSRNOT00000009127
|
Adamtsl1
|
ADAMTS-like 1 |
| chr5_+_151692108 | 29.51 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr20_-_4879779 | 29.35 |
ENSRNOT00000081924
|
Hspa1b
|
heat shock protein family A (Hsp70) member 1B |
| chr2_-_197860699 | 29.32 |
ENSRNOT00000028740
|
Ecm1
|
extracellular matrix protein 1 |
| chr16_+_6962722 | 29.15 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr1_+_137799185 | 29.13 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr19_+_27404712 | 29.00 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr1_-_215033460 | 28.69 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr5_-_64850427 | 28.35 |
ENSRNOT00000008883
|
Tmem246
|
transmembrane protein 246 |
| chr19_-_9777465 | 28.05 |
ENSRNOT00000017413
|
Ndrg4
|
NDRG family member 4 |
| chr17_-_13393243 | 28.00 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr18_-_15540177 | 27.85 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr10_+_47765432 | 27.65 |
ENSRNOT00000078231
|
LOC102553715
|
microfibril-associated glycoprotein 4-like |
| chr16_+_54332660 | 27.46 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr2_+_60920257 | 27.30 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr18_-_57201740 | 26.78 |
ENSRNOT00000026227
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr2_-_231521052 | 26.37 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr7_+_2752680 | 26.34 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr2_-_180914940 | 26.25 |
ENSRNOT00000015732
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr13_+_52662996 | 25.99 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr1_+_199941161 | 25.88 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr8_+_119228612 | 25.83 |
ENSRNOT00000078439
ENSRNOT00000043737 |
Lrrc2
|
leucine rich repeat containing 2 |
| chr4_-_112357484 | 25.38 |
ENSRNOT00000057854
|
AABR07061254.1
|
|
| chr20_-_2701815 | 25.17 |
ENSRNOT00000061950
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr14_-_13058172 | 24.86 |
ENSRNOT00000002746
ENSRNOT00000071706 |
Prdm8
|
PR/SET domain 8 |
| chr19_+_25095089 | 24.72 |
ENSRNOT00000041717
|
Prkaca
|
protein kinase cAMP-activated catalytic subunit alpha |
| chr4_+_44597123 | 23.99 |
ENSRNOT00000078250
|
Cav1
|
caveolin 1 |
| chr6_-_108076186 | 23.94 |
ENSRNOT00000014814
|
Fam161b
|
family with sequence similarity 161, member B |
| chr15_+_102164751 | 23.61 |
ENSRNOT00000076400
|
Gpc6
|
glypican 6 |
| chr20_+_6923489 | 23.43 |
ENSRNOT00000093373
ENSRNOT00000000632 |
Pi16
|
peptidase inhibitor 16 |
| chr6_+_108076306 | 23.31 |
ENSRNOT00000014913
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr1_-_170404056 | 23.29 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr13_-_105140473 | 23.24 |
ENSRNOT00000087023
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr1_-_275882444 | 23.24 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr10_+_63659085 | 23.10 |
ENSRNOT00000005039
|
Rilp
|
Rab interacting lysosomal protein |
| chr3_+_72540538 | 23.10 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr2_+_11658568 | 23.04 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr4_+_71836677 | 22.53 |
ENSRNOT00000024048
|
Tas2r126
|
taste receptor, type 2, member 126 |
| chr2_-_187863349 | 22.18 |
ENSRNOT00000084455
|
Lmna
|
lamin A/C |
| chr1_-_275906686 | 22.18 |
ENSRNOT00000092170
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr14_+_38247791 | 22.17 |
ENSRNOT00000064128
|
Corin
|
corin, serine peptidase |
| chr9_-_55256340 | 22.07 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr4_-_163214678 | 21.89 |
ENSRNOT00000091602
|
Clec1a
|
C-type lectin domain family 1, member A |
| chr4_+_56625561 | 21.88 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr1_-_215536770 | 21.86 |
ENSRNOT00000078030
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr3_+_140024043 | 21.84 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr20_-_45053640 | 21.81 |
ENSRNOT00000072256
|
RGD1561777
|
similar to Na+ dependent glucose transporter 1 |
| chr7_-_135630654 | 21.74 |
ENSRNOT00000047388
ENSRNOT00000088223 ENSRNOT00000074793 |
Adamts20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr2_+_222792999 | 21.39 |
ENSRNOT00000036228
|
AABR07013086.1
|
|
| chr7_+_144595960 | 21.35 |
ENSRNOT00000021909
|
Hoxc9
|
homeobox C9 |
| chr17_-_15566332 | 21.31 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr16_-_81880502 | 21.26 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr1_+_84304228 | 21.21 |
ENSRNOT00000024771
|
Prx
|
periaxin |
| chr3_+_160207913 | 21.15 |
ENSRNOT00000014346
|
Wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr4_-_89281222 | 21.14 |
ENSRNOT00000010898
|
Fam13a
|
family with sequence similarity 13, member A |
| chr15_+_110114148 | 21.11 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr1_-_162385575 | 20.82 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr1_+_101012822 | 20.63 |
ENSRNOT00000027809
|
Rras
|
related RAS viral (r-ras) oncogene homolog |
| chr1_+_144239020 | 20.53 |
ENSRNOT00000032106
|
Adamtsl3
|
ADAMTS-like 3 |
| chr14_-_60964324 | 20.29 |
ENSRNOT00000005155
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr5_-_139749050 | 20.21 |
ENSRNOT00000056651
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr14_+_83560541 | 20.10 |
ENSRNOT00000057738
ENSRNOT00000085228 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_-_24056373 | 20.01 |
ENSRNOT00000015566
|
Slc2a12
|
solute carrier family 2 member 12 |
| chr4_-_71893306 | 19.89 |
ENSRNOT00000077710
|
Olr803
|
olfactory receptor 803 |
| chr4_-_84768249 | 19.57 |
ENSRNOT00000013205
|
Fkbp14
|
FK506 binding protein 14 |
| chr20_+_5050327 | 19.47 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr20_+_27954433 | 19.29 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr2_+_256609787 | 19.19 |
ENSRNOT00000047564
ENSRNOT00000088437 |
Adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr8_-_107952530 | 19.19 |
ENSRNOT00000052043
|
Cldn18
|
claudin 18 |
| chr14_+_60857989 | 19.12 |
ENSRNOT00000034411
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr15_+_10120206 | 19.08 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr5_-_64789318 | 19.04 |
ENSRNOT00000078957
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr3_-_48536235 | 18.82 |
ENSRNOT00000083536
|
Fap
|
fibroblast activation protein, alpha |
| chr15_+_23665202 | 18.75 |
ENSRNOT00000089226
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr10_+_45659143 | 18.50 |
ENSRNOT00000058327
|
Wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr1_-_250626844 | 18.31 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr10_+_46972038 | 18.21 |
ENSRNOT00000023838
|
Mief2
|
mitochondrial elongation factor 2 |
| chr9_-_83253458 | 18.15 |
ENSRNOT00000041689
|
Epha4
|
Eph receptor A4 |
| chr8_-_105462141 | 18.15 |
ENSRNOT00000066731
ENSRNOT00000078760 |
Clstn2
|
calsyntenin 2 |
| chr13_-_80738634 | 18.10 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr2_-_172361779 | 18.10 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr2_-_30246010 | 18.10 |
ENSRNOT00000023900
|
Mccc2
|
methylcrotonoyl-CoA carboxylase 2 |
| chr2_+_142262236 | 17.92 |
ENSRNOT00000019057
|
Lhfp
|
lipoma HMGIC fusion partner |
| chr17_+_75190783 | 17.88 |
ENSRNOT00000085840
|
AABR07028568.1
|
|
| chr3_+_11679530 | 17.75 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr12_+_23752844 | 17.70 |
ENSRNOT00000001953
|
Ssc4d
|
scavenger receptor cysteine rich family member with 4 domains |
| chr16_+_59077574 | 17.63 |
ENSRNOT00000090477
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr13_-_80775230 | 17.62 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr7_-_143523457 | 17.56 |
ENSRNOT00000012943
ENSRNOT00000082264 |
Krt4
|
keratin 4 |
| chr10_+_62191656 | 17.56 |
ENSRNOT00000029866
|
Smyd4
|
SET and MYND domain containing 4 |
| chr19_+_61332351 | 17.55 |
ENSRNOT00000014492
|
Nrp1
|
neuropilin 1 |
| chr13_+_57131395 | 17.54 |
ENSRNOT00000017884
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr19_+_16417817 | 17.43 |
ENSRNOT00000015272
|
Irx5
|
iroquois homeobox 5 |
| chr16_+_6970342 | 17.38 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfia
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 64.5 | 258.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 50.1 | 150.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 37.8 | 151.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 29.8 | 89.4 | GO:0031439 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 27.0 | 81.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 24.0 | 72.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 22.0 | 66.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 21.0 | 147.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 17.2 | 51.5 | GO:1902304 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of potassium ion export(GO:1902304) |
| 16.8 | 201.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 16.6 | 66.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 15.4 | 184.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 15.2 | 45.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 15.1 | 75.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 14.2 | 42.7 | GO:1904057 | negative regulation of SMAD protein import into nucleus(GO:0060392) negative regulation of sensory perception of pain(GO:1904057) |
| 14.0 | 139.5 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 13.5 | 53.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 13.0 | 65.0 | GO:0098907 | regulation of SA node cell action potential(GO:0098907) |
| 12.4 | 37.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 12.3 | 36.8 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 12.1 | 60.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 12.0 | 36.0 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 12.0 | 24.0 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 10.7 | 74.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 10.6 | 31.9 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 10.4 | 31.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 10.4 | 51.8 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 10.0 | 40.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 10.0 | 40.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 9.8 | 9.8 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 9.2 | 46.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 9.1 | 118.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 9.1 | 72.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 8.9 | 35.4 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 7.9 | 39.7 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 7.7 | 23.0 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 7.4 | 22.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 7.3 | 36.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 7.3 | 29.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 7.2 | 50.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 7.1 | 78.2 | GO:0046085 | purine nucleoside monophosphate catabolic process(GO:0009128) adenosine metabolic process(GO:0046085) |
| 7.1 | 21.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 6.9 | 20.7 | GO:0097187 | dentinogenesis(GO:0097187) |
| 6.6 | 46.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 6.6 | 19.8 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 6.5 | 13.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 6.4 | 19.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 6.3 | 18.8 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 6.3 | 31.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 6.2 | 31.2 | GO:1904008 | response to monosodium glutamate(GO:1904008) |
| 6.2 | 55.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 6.2 | 67.8 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 6.0 | 18.1 | GO:2001106 | fasciculation of motor neuron axon(GO:0097156) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 6.0 | 30.2 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 6.0 | 18.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 5.9 | 17.6 | GO:0061441 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 5.4 | 16.1 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 5.1 | 15.2 | GO:0048749 | compound eye development(GO:0048749) |
| 5.0 | 45.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 4.9 | 14.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 4.9 | 14.7 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 4.9 | 29.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 4.7 | 9.4 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) kidney smooth muscle tissue development(GO:0072194) |
| 4.5 | 13.6 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 4.4 | 53.3 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 4.4 | 17.6 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 4.4 | 13.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 4.1 | 12.4 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 4.1 | 24.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 4.1 | 16.4 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 4.0 | 12.0 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 3.8 | 26.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 3.7 | 14.9 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 3.6 | 21.7 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 3.6 | 61.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 3.5 | 10.6 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 3.5 | 7.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 3.4 | 13.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 3.4 | 27.3 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 3.4 | 10.2 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 3.4 | 43.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 3.4 | 16.8 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 3.3 | 23.4 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 3.3 | 10.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 3.3 | 16.7 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 3.3 | 23.3 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 3.3 | 9.9 | GO:1903699 | tarsal gland development(GO:1903699) |
| 3.2 | 32.4 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 3.2 | 9.6 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 3.2 | 9.5 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 3.1 | 115.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 3.0 | 6.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 3.0 | 33.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 3.0 | 32.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 3.0 | 3.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 3.0 | 32.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 2.9 | 58.6 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 2.9 | 8.8 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 2.9 | 8.7 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 2.9 | 54.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 2.7 | 8.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 2.6 | 18.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 2.6 | 10.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 2.5 | 10.1 | GO:1990375 | baculum development(GO:1990375) |
| 2.5 | 25.3 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 2.5 | 12.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 2.4 | 12.2 | GO:1905050 | positive regulation of metallopeptidase activity(GO:1905050) |
| 2.4 | 19.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 2.4 | 21.4 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 2.3 | 11.5 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 2.3 | 20.6 | GO:0044351 | macropinocytosis(GO:0044351) |
| 2.3 | 29.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 2.2 | 13.3 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 2.2 | 13.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 2.2 | 6.6 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 2.2 | 6.6 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 2.1 | 6.4 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) |
| 2.0 | 18.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 2.0 | 31.9 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 2.0 | 5.9 | GO:0006296 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 1.9 | 11.7 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 1.9 | 74.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 1.9 | 9.5 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 1.9 | 7.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 1.9 | 9.4 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.9 | 42.9 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 1.9 | 46.4 | GO:0097435 | fibril organization(GO:0097435) |
| 1.8 | 9.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.8 | 34.3 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 1.8 | 5.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.8 | 19.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 1.8 | 54.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 1.7 | 28.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.7 | 12.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.7 | 8.3 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 1.6 | 8.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 1.6 | 13.0 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 1.6 | 16.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.6 | 8.0 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 1.6 | 29.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 1.5 | 9.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.5 | 29.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 1.4 | 15.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 1.4 | 7.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 1.4 | 16.6 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 1.4 | 12.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 1.3 | 21.4 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 1.3 | 1.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 1.3 | 14.7 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 1.3 | 4.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 1.3 | 9.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.3 | 20.8 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 1.3 | 5.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 1.3 | 6.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.3 | 17.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 1.2 | 3.7 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 1.2 | 10.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 1.1 | 12.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.1 | 53.4 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 1.1 | 3.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 1.1 | 12.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.1 | 27.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 1.1 | 5.5 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 1.1 | 20.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 1.1 | 18.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 1.1 | 4.3 | GO:0010916 | regulation of phosphatidylcholine catabolic process(GO:0010899) regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 1.0 | 54.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 1.0 | 3.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 1.0 | 2.0 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.9 | 8.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.9 | 8.3 | GO:0051195 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.9 | 8.9 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.9 | 14.9 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.9 | 6.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.8 | 23.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.8 | 80.0 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.8 | 18.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.8 | 2.4 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.8 | 8.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.8 | 2.4 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.8 | 9.4 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.8 | 29.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.8 | 6.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.8 | 4.6 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) negative regulation by host of viral genome replication(GO:0044828) |
| 0.8 | 48.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.7 | 1.5 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.7 | 8.1 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.7 | 5.8 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.7 | 14.6 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.7 | 9.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.7 | 10.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.7 | 4.8 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.7 | 10.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.7 | 2.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.7 | 37.0 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.7 | 3.3 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.7 | 7.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.6 | 1.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.6 | 5.1 | GO:0003416 | endochondral bone growth(GO:0003416) bone growth(GO:0098868) |
| 0.6 | 4.5 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.6 | 12.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.6 | 20.1 | GO:0048662 | negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.6 | 6.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 11.0 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.5 | 8.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.5 | 15.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.5 | 19.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.5 | 39.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.5 | 4.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.5 | 3.3 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.4 | 3.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.4 | 4.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.4 | 22.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.4 | 21.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.4 | 12.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.4 | 12.3 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.4 | 1.9 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.4 | 7.0 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.4 | 1.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.4 | 468.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.4 | 1.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.3 | 2.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 8.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 4.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 14.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.3 | 1.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.3 | 3.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.3 | 3.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.3 | 22.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.3 | 3.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.3 | 35.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.3 | 12.3 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.3 | 4.5 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.3 | 0.6 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.2 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.3 | 3.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 5.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 0.3 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.3 | 2.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.3 | 24.7 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.3 | 17.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.3 | 6.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 5.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.3 | 7.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.3 | 24.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.3 | 1.0 | GO:0071872 | cellular response to epinephrine stimulus(GO:0071872) |
| 0.2 | 20.6 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.2 | 0.7 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 2.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 4.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.2 | 2.7 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.2 | 3.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 1.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.2 | 1.6 | GO:0061548 | ganglion development(GO:0061548) |
| 0.2 | 1.9 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 0.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.2 | 5.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 0.8 | GO:0006842 | tricarboxylic acid transport(GO:0006842) |
| 0.1 | 13.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 2.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 12.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 9.4 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.1 | 4.0 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 4.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 25.1 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.1 | 16.4 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 2.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.6 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.1 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 4.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.4 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.0 | 1.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.5 | GO:0042220 | response to cocaine(GO:0042220) |
| 0.0 | 2.2 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 0.2 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.5 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.0 | 1.0 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 2.5 | GO:2000278 | regulation of DNA biosynthetic process(GO:2000278) |
| 0.0 | 1.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 2.4 | GO:0006664 | glycolipid metabolic process(GO:0006664) |
| 0.0 | 0.3 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 36.0 | 144.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 22.6 | 67.8 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 18.5 | 166.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 18.4 | 147.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 13.9 | 55.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 12.6 | 201.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 12.2 | 97.7 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 10.9 | 98.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 9.9 | 29.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 7.3 | 29.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 7.2 | 93.7 | GO:0032982 | myosin filament(GO:0032982) |
| 6.7 | 33.4 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 6.4 | 51.0 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 6.4 | 19.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 6.0 | 71.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 5.6 | 889.8 | GO:0031674 | I band(GO:0031674) |
| 5.4 | 75.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 5.4 | 32.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 5.1 | 30.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 5.0 | 60.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 4.6 | 37.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 4.6 | 36.8 | GO:0005638 | lamin filament(GO:0005638) |
| 4.5 | 31.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 4.1 | 12.3 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 3.3 | 40.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 3.3 | 16.7 | GO:1903768 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 3.3 | 16.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 3.2 | 34.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 3.0 | 24.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 2.9 | 8.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 2.9 | 38.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 2.9 | 20.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 2.7 | 24.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 2.6 | 39.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 2.6 | 12.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 2.6 | 46.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 2.3 | 97.2 | GO:0016459 | myosin complex(GO:0016459) |
| 2.2 | 15.6 | GO:0001652 | granular component(GO:0001652) |
| 2.2 | 4.5 | GO:0036396 | MIS complex(GO:0036396) |
| 2.2 | 17.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 2.2 | 30.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 2.1 | 10.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 2.1 | 6.4 | GO:0005712 | chiasma(GO:0005712) recombination nodule(GO:0005713) |
| 2.1 | 10.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 2.0 | 53.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 2.0 | 5.9 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 1.9 | 72.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 1.8 | 14.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 1.8 | 9.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.7 | 6.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.7 | 13.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.4 | 9.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.3 | 12.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 1.2 | 7.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 1.2 | 7.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 1.1 | 33.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 1.1 | 16.8 | GO:0005605 | basal lamina(GO:0005605) |
| 1.1 | 46.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 1.1 | 63.9 | GO:0030017 | sarcomere(GO:0030017) |
| 1.1 | 53.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 1.1 | 21.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.0 | 10.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.0 | 2.9 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.9 | 46.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.7 | 1.5 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.7 | 34.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.7 | 12.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.7 | 117.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.7 | 34.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.7 | 20.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.7 | 4.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.7 | 47.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.7 | 3.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.7 | 35.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.6 | 137.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.6 | 9.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.6 | 33.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.6 | 4.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.5 | 7.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.5 | 13.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.4 | 4.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.4 | 4.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 13.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.4 | 11.7 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.4 | 137.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.4 | 9.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 29.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.3 | 23.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 7.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 21.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 1.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 42.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.3 | 1.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 7.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 1.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 18.5 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 4.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 41.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 1.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 21.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 56.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.2 | 13.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 13.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 10.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 12.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 1.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 14.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 25.8 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 4.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 6.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 8.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 53.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 2.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.0 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 19.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 64.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 242.8 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.7 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 12.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 3.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 50.7 | 203.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 50.1 | 150.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 42.9 | 214.7 | GO:0051373 | FATZ binding(GO:0051373) |
| 29.8 | 89.4 | GO:0031249 | denatured protein binding(GO:0031249) |
| 15.3 | 61.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 14.8 | 59.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 14.4 | 72.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 12.4 | 37.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 11.5 | 34.4 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 10.5 | 147.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 9.8 | 48.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 9.7 | 38.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 9.3 | 37.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 8.9 | 26.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 7.6 | 30.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 7.6 | 45.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 6.7 | 53.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 6.7 | 201.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 6.6 | 19.8 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 6.5 | 26.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 6.5 | 19.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 6.2 | 24.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 5.9 | 29.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 5.5 | 60.4 | GO:0031432 | titin binding(GO:0031432) |
| 5.4 | 65.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 5.4 | 81.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 5.3 | 26.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 5.0 | 105.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 4.6 | 78.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 4.5 | 13.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 4.5 | 36.0 | GO:0043199 | sulfate binding(GO:0043199) |
| 4.5 | 13.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 4.4 | 13.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 4.3 | 55.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 4.3 | 93.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 4.2 | 50.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 4.1 | 12.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 4.1 | 16.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 4.1 | 20.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 4.0 | 12.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 3.9 | 11.8 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 3.8 | 68.9 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 3.8 | 45.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 3.8 | 45.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 3.7 | 86.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 3.7 | 33.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 3.7 | 29.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 3.6 | 18.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 3.5 | 17.7 | GO:0005534 | galactose binding(GO:0005534) |
| 3.4 | 10.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 3.3 | 16.7 | GO:0033041 | sweet taste receptor activity(GO:0033041) |
| 3.3 | 32.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 3.2 | 70.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 3.1 | 12.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 3.1 | 9.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 3.1 | 18.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 3.0 | 87.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 3.0 | 65.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 2.9 | 31.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 2.8 | 24.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.7 | 16.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 2.7 | 10.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 2.7 | 16.3 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 2.7 | 16.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 2.7 | 8.0 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 2.6 | 7.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 2.6 | 39.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 2.5 | 10.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 2.4 | 12.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 2.3 | 27.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 2.3 | 23.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 2.3 | 23.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 2.3 | 11.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 2.3 | 18.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 2.2 | 11.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 2.2 | 17.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) vascular endothelial growth factor binding(GO:0038085) |
| 2.1 | 10.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 2.1 | 6.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 2.1 | 51.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 2.0 | 6.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 2.0 | 27.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 2.0 | 27.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 1.9 | 43.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 1.9 | 22.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 1.8 | 111.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 1.8 | 30.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 1.8 | 7.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 1.7 | 8.5 | GO:0005550 | pheromone binding(GO:0005550) |
| 1.7 | 15.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.6 | 6.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 1.6 | 35.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 1.5 | 116.8 | GO:0005518 | collagen binding(GO:0005518) |
| 1.5 | 19.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.5 | 4.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 1.5 | 31.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.5 | 5.9 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 1.4 | 44.8 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 1.4 | 9.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.3 | 46.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 1.3 | 9.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.3 | 5.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 1.3 | 6.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 1.3 | 19.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 1.3 | 8.8 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.3 | 17.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 1.2 | 46.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.2 | 14.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 1.2 | 13.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 1.2 | 14.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 1.2 | 4.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.1 | 29.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 1.1 | 9.1 | GO:0008430 | selenium binding(GO:0008430) |
| 1.1 | 8.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 1.1 | 37.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 1.1 | 29.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.1 | 3.2 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.1 | 64.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.0 | 28.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 1.0 | 31.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 1.0 | 3.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.9 | 10.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.9 | 8.5 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.9 | 14.9 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.9 | 60.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.9 | 17.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.9 | 26.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.9 | 21.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.8 | 12.7 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.8 | 4.8 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.8 | 9.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.8 | 8.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.8 | 9.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.7 | 18.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.7 | 2.9 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.7 | 11.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.7 | 3.3 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.6 | 6.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 26.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 3.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.6 | 12.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.6 | 205.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.6 | 8.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 6.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.5 | 8.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.5 | 4.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.5 | 2.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.5 | 26.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.5 | 17.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.5 | 11.9 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) |
| 0.5 | 3.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 10.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 2.7 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 4.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.4 | 59.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.4 | 9.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 2.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 6.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.4 | 5.5 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.4 | 2.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 7.5 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.4 | 9.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.4 | 1.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 468.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.4 | 224.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.4 | 3.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 6.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 1.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.3 | 7.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 4.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 1.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 7.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 0.7 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 3.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 11.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.2 | 32.0 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.2 | 1.9 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 2.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 4.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 14.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 11.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.2 | 14.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 0.8 | GO:0015142 | tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 25.0 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.1 | 14.6 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 7.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 11.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 6.3 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 4.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 30.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 3.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 5.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 0.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.5 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of synapse(GO:0098918) structural constituent of postsynaptic density(GO:0098919) |
| 0.0 | 2.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 31.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 4.2 | 188.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 3.3 | 165.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 2.8 | 30.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 2.7 | 43.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 2.4 | 9.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 2.2 | 103.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 2.1 | 39.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 1.8 | 306.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 1.7 | 33.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 1.5 | 20.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 1.4 | 8.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 1.4 | 58.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.3 | 12.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 1.2 | 28.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 1.2 | 40.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 1.2 | 30.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 1.1 | 12.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 1.1 | 64.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 1.0 | 11.5 | PID MYC PATHWAY | C-MYC pathway |
| 1.0 | 15.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 1.0 | 34.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.9 | 16.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.9 | 130.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.9 | 37.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.9 | 30.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.8 | 18.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.8 | 29.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.8 | 18.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.8 | 26.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.8 | 12.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.8 | 25.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.8 | 42.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.7 | 14.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.7 | 7.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.7 | 9.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.7 | 14.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.7 | 13.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.6 | 30.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.6 | 11.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.6 | 2.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.6 | 131.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.5 | 16.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.5 | 42.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.5 | 19.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 3.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 12.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.4 | 4.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.4 | 8.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 8.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 5.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 9.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 7.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 10.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 10.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 2.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 1.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 7.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.6 | 492.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 7.9 | 79.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 5.6 | 222.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 5.2 | 46.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 4.2 | 58.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 3.8 | 11.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 3.3 | 86.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 3.1 | 72.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 3.0 | 26.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.9 | 123.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 2.9 | 51.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 2.4 | 38.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 2.4 | 45.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 2.1 | 21.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.8 | 24.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.8 | 85.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 1.8 | 40.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 1.7 | 20.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.6 | 28.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 1.6 | 29.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 1.5 | 32.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.5 | 23.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 1.4 | 70.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 1.3 | 39.3 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 1.2 | 24.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.0 | 10.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.9 | 11.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.9 | 13.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.9 | 8.7 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.8 | 9.1 | REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | Genes involved in MAPK targets/ Nuclear events mediated by MAP kinases |
| 0.7 | 12.7 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.7 | 6.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.7 | 48.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.7 | 15.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.7 | 8.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.6 | 8.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.6 | 12.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.6 | 12.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.6 | 19.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 7.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.5 | 29.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.5 | 4.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.5 | 14.3 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.5 | 13.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.5 | 88.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.5 | 27.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.5 | 22.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.5 | 7.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 3.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 10.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 8.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.4 | 8.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 12.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.3 | 13.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 11.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 6.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 14.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 8.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 2.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 5.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.2 | 11.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.5 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 8.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 3.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 9.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |