Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
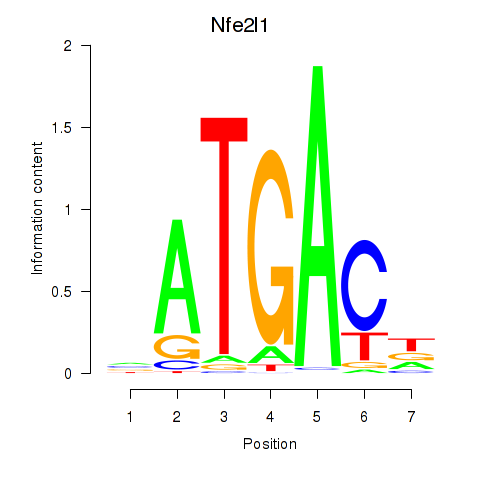
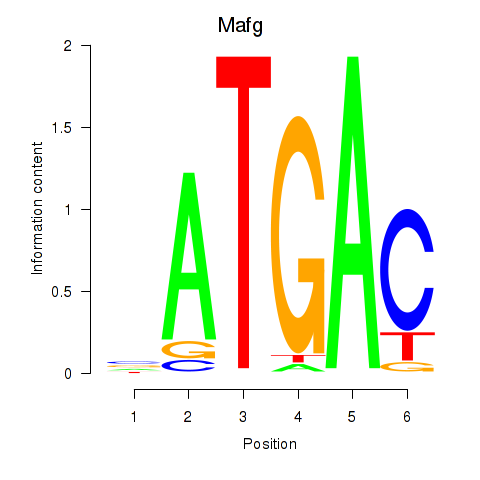
Results for Nfe2l1_Mafg
Z-value: 0.89
Transcription factors associated with Nfe2l1_Mafg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l1
|
ENSRNOG00000008830 | nuclear factor, erythroid 2-like 1 |
|
Mafg
|
ENSRNOG00000036697 | MAF bZIP transcription factor G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2l1 | rn6_v1_chr10_-_84698886_84698886 | 0.34 | 2.9e-10 | Click! |
| Mafg | rn6_v1_chr10_-_109811323_109811323 | -0.26 | 2.9e-06 | Click! |
Activity profile of Nfe2l1_Mafg motif
Sorted Z-values of Nfe2l1_Mafg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_70687500 | 32.98 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr9_-_115850634 | 32.82 |
ENSRNOT00000049720
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr13_-_103080920 | 25.27 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr18_-_63825408 | 25.25 |
ENSRNOT00000043709
|
RGD1562758
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr11_-_72109964 | 23.26 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr3_+_57770948 | 20.88 |
ENSRNOT00000038429
|
AC107446.2
|
|
| chr10_-_102289837 | 19.81 |
ENSRNOT00000044922
|
AABR07030729.1
|
|
| chr1_+_193537137 | 19.72 |
ENSRNOT00000029967
|
AABR07005667.1
|
|
| chr3_+_132052612 | 16.86 |
ENSRNOT00000030148
|
AABR07053879.1
|
|
| chr2_+_222792999 | 16.62 |
ENSRNOT00000036228
|
AABR07013086.1
|
|
| chr5_+_115649046 | 15.87 |
ENSRNOT00000041328
|
LOC108351137
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr13_+_56262190 | 15.63 |
ENSRNOT00000032908
|
AABR07021086.1
|
|
| chr3_-_46726946 | 15.54 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr16_+_25773602 | 13.91 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr16_+_16949232 | 12.68 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr8_+_100260049 | 9.58 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr8_-_36410612 | 9.14 |
ENSRNOT00000091308
|
Foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr6_+_55085313 | 8.07 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chrM_+_7006 | 8.00 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr3_-_2453933 | 7.86 |
ENSRNOT00000014060
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr1_+_229107911 | 7.18 |
ENSRNOT00000016484
|
Olr338
|
olfactory receptor 338 |
| chr6_-_108076186 | 7.13 |
ENSRNOT00000014814
|
Fam161b
|
family with sequence similarity 161, member B |
| chr1_+_106998623 | 6.70 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr3_-_51054378 | 6.46 |
ENSRNOT00000089243
|
Grb14
|
growth factor receptor bound protein 14 |
| chr7_-_117259791 | 6.36 |
ENSRNOT00000086550
|
Plec
|
plectin |
| chr1_-_222350173 | 6.27 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr1_-_215536980 | 5.95 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr10_-_59984862 | 5.90 |
ENSRNOT00000087700
|
Olr1485
|
olfactory receptor 1485 |
| chr14_-_77222006 | 5.53 |
ENSRNOT00000007074
|
Drd5
|
dopamine receptor D5 |
| chr1_-_215536770 | 5.29 |
ENSRNOT00000078030
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr10_-_78690857 | 5.23 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr8_+_36410683 | 5.10 |
ENSRNOT00000015177
|
Srpra
|
SRP receptor alpha subunit |
| chr11_-_83483513 | 4.60 |
ENSRNOT00000084380
|
AABR07034673.1
|
|
| chr2_-_77784522 | 4.52 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr4_-_161757447 | 4.48 |
ENSRNOT00000008737
|
Fkbp4
|
FK506 binding protein 4 |
| chr6_+_108076306 | 4.25 |
ENSRNOT00000014913
|
Coq6
|
coenzyme Q6 monooxygenase |
| chr1_+_166991474 | 4.22 |
ENSRNOT00000027150
|
LOC100361543
|
RhoA activator C11orf59-like |
| chr10_-_60542410 | 4.21 |
ENSRNOT00000082426
|
Olr1498
|
olfactory receptor 1498 |
| chr10_-_60006037 | 4.13 |
ENSRNOT00000065541
|
Olr1485
|
olfactory receptor 1485 |
| chr19_-_15840990 | 4.11 |
ENSRNOT00000015583
|
Irx3
|
iroquois homeobox 3 |
| chr1_-_22570303 | 3.59 |
ENSRNOT00000035539
|
Taar3
|
trace amine-associated receptor 3 |
| chr6_+_10912383 | 3.55 |
ENSRNOT00000061747
ENSRNOT00000086247 |
Ttc7a
|
tetratricopeptide repeat domain 7A |
| chr3_-_78235760 | 3.32 |
ENSRNOT00000008435
|
Olr698
|
olfactory receptor 698 |
| chr8_+_43722228 | 3.15 |
ENSRNOT00000084016
|
Olr1325
|
olfactory receptor 1325 |
| chr1_+_229877560 | 3.05 |
ENSRNOT00000089953
|
Olr347
|
olfactory receptor 347 |
| chr3_+_78748001 | 2.88 |
ENSRNOT00000008715
|
Olr722
|
olfactory receptor 722 |
| chr6_-_102353403 | 2.84 |
ENSRNOT00000090407
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B |
| chr1_-_209641123 | 2.83 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr10_-_14848967 | 2.81 |
ENSRNOT00000080944
|
Sstr5
|
somatostatin receptor 5 |
| chr5_+_154800226 | 2.78 |
ENSRNOT00000016046
|
Htr1d
|
5-hydroxytryptamine receptor 1D |
| chr8_-_43249887 | 2.73 |
ENSRNOT00000060094
|
Olr1305
|
olfactory receptor 1305 |
| chr13_+_92249759 | 2.68 |
ENSRNOT00000089352
|
Olr1596
|
olfactory receptor 1596 |
| chr9_+_74124016 | 2.67 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr10_-_12571316 | 2.65 |
ENSRNOT00000074440
|
Olr1372
|
olfactory receptor 1372 |
| chr8_-_43082948 | 2.63 |
ENSRNOT00000072050
|
LOC100910667
|
olfactory receptor 8G5-like |
| chr9_-_86103158 | 2.48 |
ENSRNOT00000021528
|
Cul3
|
cullin 3 |
| chr1_-_61626454 | 2.43 |
ENSRNOT00000038190
|
Vom1r22
|
vomeronasal 1 receptor 22 |
| chr13_+_92504374 | 2.32 |
ENSRNOT00000033697
|
Olr1602
|
olfactory receptor 1602 |
| chr3_-_76926151 | 2.31 |
ENSRNOT00000049517
|
Olr641
|
olfactory receptor 641 |
| chr5_-_58917822 | 2.31 |
ENSRNOT00000082554
|
AABR07048059.1
|
|
| chr10_+_36530719 | 2.28 |
ENSRNOT00000046476
|
Olr1409
|
olfactory receptor 1409 |
| chr3_+_21313227 | 2.26 |
ENSRNOT00000036335
|
LOC690140
|
similar to olfactory receptor Olr1374 |
| chr1_-_230766515 | 2.24 |
ENSRNOT00000040049
|
Olr383
|
olfactory receptor 383 |
| chr8_+_43763859 | 2.21 |
ENSRNOT00000080504
|
Olr1328
|
olfactory receptor 1328 |
| chr6_+_28557633 | 2.18 |
ENSRNOT00000087058
|
Gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr3_+_73988945 | 2.14 |
ENSRNOT00000050404
|
Olr515
|
olfactory receptor 515 |
| chr17_+_11101306 | 2.10 |
ENSRNOT00000030893
|
Drd1
|
dopamine receptor D1 |
| chr17_-_81558565 | 2.10 |
ENSRNOT00000048715
|
AABR07028691.1
|
|
| chr15_-_29810811 | 2.09 |
ENSRNOT00000060386
|
AABR07017662.1
|
|
| chr1_+_230576298 | 2.05 |
ENSRNOT00000049799
|
Olr374
|
olfactory receptor 374 |
| chr1_+_70454322 | 1.95 |
ENSRNOT00000072272
|
Olr4
|
olfactory receptor 4 |
| chr3_+_110508403 | 1.90 |
ENSRNOT00000079959
|
AABR07053500.2
|
|
| chr10_-_88000423 | 1.90 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr3_-_73755510 | 1.83 |
ENSRNOT00000083628
|
Olr502
|
olfactory receptor 502 |
| chr19_+_28377249 | 1.79 |
ENSRNOT00000059668
|
Vom1r21
|
vomeronasal 1 receptor 21 |
| chr1_+_229480184 | 1.78 |
ENSRNOT00000044115
|
Olr387
|
olfactory receptor 387 |
| chr8_-_43235208 | 1.73 |
ENSRNOT00000060095
|
Olr1304
|
olfactory receptor 1304 |
| chr2_+_62360754 | 1.63 |
ENSRNOT00000017276
|
Golph3
|
golgi phosphoprotein 3 |
| chr9_+_74960255 | 1.53 |
ENSRNOT00000077342
|
AABR07067976.1
|
|
| chr1_+_167748100 | 1.51 |
ENSRNOT00000024961
|
Olr45
|
olfactory receptor 45 |
| chr3_-_102773801 | 1.49 |
ENSRNOT00000047051
|
Olr767
|
olfactory receptor 767 |
| chr1_-_169973241 | 1.46 |
ENSRNOT00000023315
|
Olr191-ps
|
olfactory receptor pseudogene 191 |
| chrX_+_133227660 | 1.42 |
ENSRNOT00000004591
|
Actrt1
|
actin-related protein T1 |
| chr3_-_72818843 | 1.40 |
ENSRNOT00000041926
|
Olr442
|
olfactory receptor 442 |
| chr1_+_22319353 | 1.39 |
ENSRNOT00000038523
|
Taar9
|
trace amine-associated receptor 9 |
| chr16_-_54899347 | 1.36 |
ENSRNOT00000016321
|
Vps37a
|
VPS37A, ESCRT-I subunit |
| chr17_-_45688058 | 1.31 |
ENSRNOT00000086807
|
Olr1662
|
olfactory receptor 1662 |
| chr15_-_28446785 | 1.30 |
ENSRNOT00000015439
|
Olr1638
|
olfactory receptor 1638 |
| chr17_+_36771639 | 1.26 |
ENSRNOT00000015147
|
Gpr50
|
G protein-coupled receptor 50 |
| chr1_+_169552487 | 1.17 |
ENSRNOT00000023119
|
Olr152
|
olfactory receptor 152 |
| chr10_-_12731897 | 1.15 |
ENSRNOT00000042582
|
Olr1378
|
olfactory receptor 1378 |
| chr1_+_70480941 | 1.15 |
ENSRNOT00000072404
|
Olr6
|
olfactory receptor 6 |
| chr9_+_50966766 | 1.13 |
ENSRNOT00000076636
|
Ercc5
|
ERCC excision repair 5, endonuclease |
| chr3_+_16039249 | 1.10 |
ENSRNOT00000046196
|
Olr403
|
olfactory receptor 403 |
| chr7_-_81592206 | 1.07 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr7_-_7311272 | 1.06 |
ENSRNOT00000040812
|
Olr1020
|
olfactory receptor 1020 |
| chr1_+_171274305 | 1.04 |
ENSRNOT00000051038
|
Olr234
|
olfactory receptor 234 |
| chr1_+_230217215 | 1.02 |
ENSRNOT00000072772
|
LOC687088
|
similar to olfactory receptor 1467 |
| chr8_+_43382176 | 1.01 |
ENSRNOT00000047759
|
Olr1311
|
olfactory receptor 1311 |
| chr2_-_89498395 | 0.99 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr1_-_22559830 | 0.97 |
ENSRNOT00000047810
|
Taar4
|
trace amine-associated receptor 4 |
| chr7_+_5401508 | 0.88 |
ENSRNOT00000050898
|
Olr903
|
olfactory receptor 903 |
| chr1_+_230450529 | 0.86 |
ENSRNOT00000045383
|
Olr371
|
olfactory receptor 371 |
| chr15_+_27243268 | 0.74 |
ENSRNOT00000046583
|
Olr1615
|
olfactory receptor 1615 |
| chr10_-_87248572 | 0.74 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr11_+_65610352 | 0.69 |
ENSRNOT00000065766
|
LOC688739
|
similar to Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) |
| chr1_+_212181374 | 0.60 |
ENSRNOT00000085921
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr11_-_32469566 | 0.56 |
ENSRNOT00000002718
|
LOC498063
|
similar to RIKEN cDNA 4930563D23 |
| chr5_-_137515690 | 0.53 |
ENSRNOT00000049593
|
LOC100912421
|
olfactory receptor 2D2-like |
| chr4_+_72670543 | 0.52 |
ENSRNOT00000044838
|
Olr821
|
olfactory receptor 821 |
| chr16_+_54899452 | 0.39 |
ENSRNOT00000016882
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr10_-_47825252 | 0.31 |
ENSRNOT00000087102
|
Epn2
|
epsin 2 |
| chr9_+_95221474 | 0.25 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr3_-_20853708 | 0.23 |
ENSRNOT00000010360
|
Olr411
|
olfactory receptor 411 |
| chrX_-_49530166 | 0.19 |
ENSRNOT00000072026
|
Srsf8
|
serine and arginine rich splicing factor 8 |
| chr8_-_40564556 | 0.15 |
ENSRNOT00000075779
|
Olfr883
|
olfactory receptor 883 |
| chr2_-_252451999 | 0.13 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr3_+_114236718 | 0.13 |
ENSRNOT00000024201
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr1_+_230377079 | 0.12 |
ENSRNOT00000042937
|
Olr367
|
olfactory receptor 367 |
| chr17_-_45746753 | 0.05 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l1_Mafg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.1 | 15.5 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 1.8 | 5.5 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 1.3 | 6.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 1.1 | 7.9 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.0 | 4.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.8 | 4.2 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.8 | 6.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.6 | 2.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.5 | 1.6 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.5 | 2.7 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.5 | 2.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.5 | 2.8 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.5 | 9.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.4 | 5.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 2.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.4 | 1.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 6.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 2.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 4.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 4.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 1.8 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 8.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 6.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 3.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 72.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.0 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.4 | GO:0043928 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 7.8 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 15.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.9 | 5.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.6 | 4.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.5 | 8.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 2.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.4 | 6.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 12.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 6.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 1.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.2 | 9.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 2.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 4.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 2.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 6.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 71.0 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 15.9 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.5 | 7.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.6 | 7.9 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.9 | 4.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.9 | 5.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.8 | 2.5 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.5 | 2.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.4 | 6.7 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.3 | 5.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 8.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 1.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 6.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 2.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 2.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 6.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 2.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.2 | 4.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 6.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 1.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 5.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 12.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 27.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 1.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 4.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 45.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 4.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 15.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 7.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 6.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 15.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 6.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 14.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |