Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
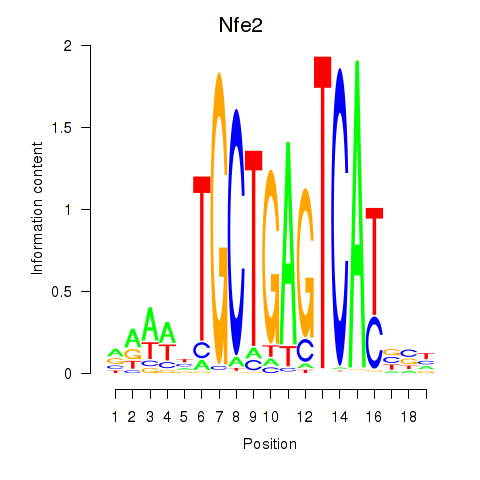
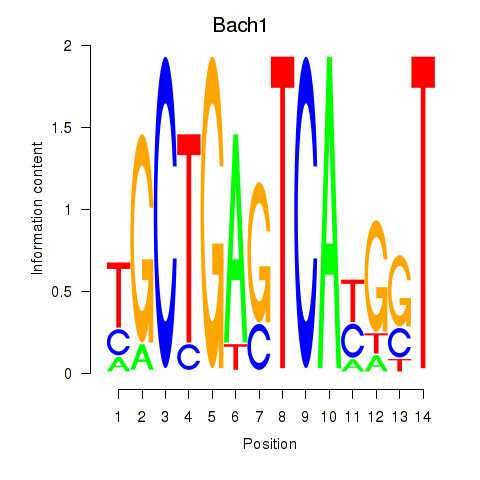
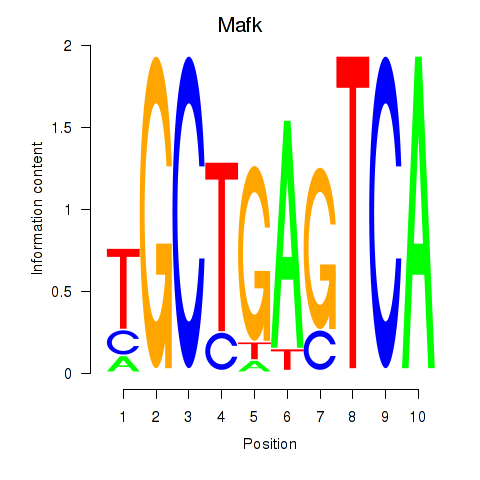
Results for Nfe2_Bach1_Mafk
Z-value: 1.23
Transcription factors associated with Nfe2_Bach1_Mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2
|
ENSRNOG00000036837 | nuclear factor, erythroid 2 |
|
Bach1
|
ENSRNOG00000001582 | BTB domain and CNC homolog 1 |
|
Mafk
|
ENSRNOG00000001277 | MAF bZIP transcription factor K |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2 | rn6_v1_chr7_-_144880092_144880092 | -0.56 | 2.0e-27 | Click! |
| Bach1 | rn6_v1_chr11_+_27364916_27364916 | -0.52 | 4.2e-23 | Click! |
| Mafk | rn6_v1_chr12_-_16934706_16934706 | -0.29 | 9.3e-08 | Click! |
Activity profile of Nfe2_Bach1_Mafk motif
Sorted Z-values of Nfe2_Bach1_Mafk motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_64929721 | 59.32 |
ENSRNOT00000068633
|
Aox1
|
aldehyde oxidase 1 |
| chr19_+_52077109 | 59.17 |
ENSRNOT00000020225
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr1_-_198232344 | 58.10 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr19_+_52077501 | 54.74 |
ENSRNOT00000079240
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr8_-_94368834 | 49.74 |
ENSRNOT00000078977
|
Me1
|
malic enzyme 1 |
| chr3_-_10371240 | 48.20 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr4_+_163293724 | 41.54 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr14_+_2050483 | 40.94 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chrX_-_31968152 | 39.27 |
ENSRNOT00000004884
|
Pir
|
pirin |
| chr19_+_38422164 | 31.76 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr11_+_30363280 | 28.98 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chrX_-_54303729 | 28.34 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr1_+_199248470 | 27.21 |
ENSRNOT00000025933
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr14_-_115052450 | 26.07 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr1_-_221917901 | 26.01 |
ENSRNOT00000092270
|
Slc22a12
|
solute carrier family 22 member 12 |
| chr5_+_135536413 | 25.62 |
ENSRNOT00000023132
|
Prdx1
|
peroxiredoxin 1 |
| chr15_+_2733114 | 24.88 |
ENSRNOT00000074545
|
LOC108348179
|
dual specificity protein phosphatase 13 |
| chr2_+_225827504 | 24.73 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr18_-_27749235 | 23.01 |
ENSRNOT00000026696
|
Hspa9
|
heat shock protein family A member 9 |
| chrX_+_17171605 | 22.84 |
ENSRNOT00000048236
|
Nudt10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr4_+_30807879 | 21.75 |
ENSRNOT00000013184
|
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr7_+_59200918 | 21.60 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr15_+_2526368 | 20.83 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chr19_-_64303 | 20.30 |
ENSRNOT00000015451
|
Ces2a
|
carboxylesterase 2A |
| chr8_-_130550388 | 19.13 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr10_-_35716260 | 19.12 |
ENSRNOT00000004308
ENSRNOT00000059255 |
Sqstm1
|
sequestosome 1 |
| chr16_-_14382641 | 18.61 |
ENSRNOT00000018723
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr13_-_99287887 | 16.02 |
ENSRNOT00000004780
|
Ephx1
|
epoxide hydrolase 1 |
| chr1_+_282568287 | 16.00 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr4_-_115332052 | 15.51 |
ENSRNOT00000017643
|
Clec4f
|
C-type lectin domain family 4, member F |
| chr6_-_129010271 | 14.99 |
ENSRNOT00000075378
|
Serpina10
|
serpin family A member 10 |
| chr6_+_42092467 | 14.99 |
ENSRNOT00000060499
|
E2f6
|
E2F transcription factor 6 |
| chr1_-_166943592 | 14.98 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr8_+_42613601 | 14.53 |
ENSRNOT00000040297
|
Olr1264
|
olfactory receptor 1264 |
| chr7_-_123630045 | 14.14 |
ENSRNOT00000050002
|
Cyp2d1
|
cytochrome P450, family 2, subfamily d, polypeptide 1 |
| chr6_+_128073344 | 13.81 |
ENSRNOT00000014073
|
LOC500712
|
Ab1-233 |
| chr13_-_36101411 | 13.80 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr8_-_73194837 | 13.38 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr1_+_277068761 | 12.93 |
ENSRNOT00000044183
ENSRNOT00000022382 |
Habp2
|
hyaluronan binding protein 2 |
| chr9_+_17817721 | 12.78 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr3_+_152552822 | 12.27 |
ENSRNOT00000089719
|
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr5_-_58445953 | 12.11 |
ENSRNOT00000046102
|
Vcp
|
valosin-containing protein |
| chr9_+_93080615 | 12.07 |
ENSRNOT00000024306
|
Psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr11_+_86797557 | 11.79 |
ENSRNOT00000083049
ENSRNOT00000046594 |
Tango2
|
transport and golgi organization 2 homolog |
| chr1_+_282567674 | 11.65 |
ENSRNOT00000090543
|
Ces2i
|
carboxylesterase 2I |
| chr8_+_43705216 | 11.63 |
ENSRNOT00000090425
|
AC115384.1
|
|
| chr9_+_10963723 | 11.54 |
ENSRNOT00000075424
|
Plin4
|
perilipin 4 |
| chr7_+_3355116 | 10.99 |
ENSRNOT00000030453
ENSRNOT00000061657 |
Itga7
|
integrin subunit alpha 7 |
| chrX_-_142248369 | 10.95 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr17_+_53102534 | 10.64 |
ENSRNOT00000021433
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr9_+_51298426 | 10.60 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_242882306 | 10.52 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr2_-_220535751 | 10.46 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chr18_-_59086251 | 10.32 |
ENSRNOT00000025510
ENSRNOT00000083831 |
Txnl1
|
thioredoxin-like 1 |
| chr6_+_64224861 | 10.23 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr9_+_119353840 | 10.07 |
ENSRNOT00000085362
|
Myom1
|
myomesin 1 |
| chr5_-_33892462 | 9.73 |
ENSRNOT00000009334
|
Atp6v0d2
|
ATPase H+ transporting V0 subunit D2 |
| chr7_-_117680004 | 9.60 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr1_+_79989019 | 9.43 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr18_+_56364620 | 9.23 |
ENSRNOT00000068535
ENSRNOT00000086033 |
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr20_-_422464 | 9.01 |
ENSRNOT00000051646
|
Olr1673
|
olfactory receptor 1673 |
| chr11_+_87435185 | 8.69 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr2_+_60180215 | 8.64 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr20_+_5008508 | 8.60 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr10_-_18574710 | 8.49 |
ENSRNOT00000079031
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr3_-_8981362 | 8.23 |
ENSRNOT00000086717
|
Crat
|
carnitine O-acetyltransferase |
| chr7_-_117254765 | 8.11 |
ENSRNOT00000006311
|
Plec
|
plectin |
| chr17_-_87826421 | 8.01 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr3_-_159802952 | 7.70 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr9_-_85243001 | 7.67 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr4_-_113902399 | 7.54 |
ENSRNOT00000087788
|
Tlx2
|
T-cell leukemia homeobox 2 |
| chr14_-_72380330 | 7.52 |
ENSRNOT00000082653
ENSRNOT00000081878 ENSRNOT00000006727 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr15_-_44860604 | 7.47 |
ENSRNOT00000018637
|
Nefm
|
neurofilament medium |
| chr14_+_107750162 | 7.35 |
ENSRNOT00000012767
|
Zrsr1
|
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 |
| chr4_-_97784842 | 7.29 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr11_+_66713888 | 7.16 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr1_+_41192824 | 7.11 |
ENSRNOT00000082133
|
Esr1
|
estrogen receptor 1 |
| chrX_-_104791210 | 6.98 |
ENSRNOT00000004921
|
Sytl4
|
synaptotagmin-like 4 |
| chr6_+_93423002 | 6.98 |
ENSRNOT00000010753
|
Psma3
|
proteasome subunit alpha 3 |
| chr1_+_193187972 | 6.98 |
ENSRNOT00000090172
ENSRNOT00000065342 |
Slc5a11
|
solute carrier family 5 member 11 |
| chr5_+_64294321 | 6.93 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr1_+_264507985 | 6.91 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chr3_-_11410732 | 6.88 |
ENSRNOT00000034930
|
RGD1561113
|
similar to Hypothetical UPF0184 protein C9orf16 homolog |
| chr8_+_70712513 | 6.81 |
ENSRNOT00000079548
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr12_+_39554171 | 6.57 |
ENSRNOT00000024347
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr17_-_53102342 | 6.49 |
ENSRNOT00000073834
|
Psma2
|
proteasome subunit alpha 2 |
| chr5_-_113578928 | 6.40 |
ENSRNOT00000010621
|
Plaa
|
phospholipase A2, activating protein |
| chr16_+_20336638 | 6.21 |
ENSRNOT00000080467
|
Kcnn1
|
potassium calcium-activated channel subfamily N member 1 |
| chr8_+_42009816 | 6.17 |
ENSRNOT00000071964
|
LOC100911801
|
olfactory receptor 143-like |
| chr16_+_23317953 | 6.08 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr11_-_76888178 | 6.05 |
ENSRNOT00000049841
|
Ostn
|
osteocrin |
| chr14_+_2892753 | 6.04 |
ENSRNOT00000061630
|
Evi5
|
ecotropic viral integration site 5 |
| chr7_-_117267402 | 5.93 |
ENSRNOT00000088945
|
Plec
|
plectin |
| chr19_-_22626653 | 5.87 |
ENSRNOT00000078348
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr19_-_39646693 | 5.79 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr16_+_10727571 | 5.72 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr10_-_16045835 | 5.57 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_+_128629942 | 5.42 |
ENSRNOT00000010462
ENSRNOT00000010440 |
Nrdc
|
nardilysin convertase |
| chr7_-_130120579 | 5.27 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr12_-_6784111 | 5.25 |
ENSRNOT00000078541
|
AABR07035203.1
|
|
| chr1_-_31881531 | 5.25 |
ENSRNOT00000081225
|
Tppp
|
tubulin polymerization promoting protein |
| chr4_-_114768029 | 5.08 |
ENSRNOT00000083404
|
Tlx2
|
T-cell leukemia homeobox 2 |
| chr7_-_117267803 | 5.08 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr1_+_167719947 | 5.08 |
ENSRNOT00000024971
|
Olr43
|
olfactory receptor 43 |
| chr15_+_8730871 | 5.01 |
ENSRNOT00000081845
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr5_+_59063531 | 4.98 |
ENSRNOT00000085311
ENSRNOT00000065909 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr2_-_187914905 | 4.97 |
ENSRNOT00000026974
|
Lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr2_+_211050360 | 4.90 |
ENSRNOT00000026928
|
Psma5
|
proteasome subunit alpha 5 |
| chr7_-_117265493 | 4.86 |
ENSRNOT00000083429
|
Plec
|
plectin |
| chr10_+_65780494 | 4.86 |
ENSRNOT00000013100
|
Poldip2
|
DNA polymerase delta interacting protein 2 |
| chr8_-_40564556 | 4.85 |
ENSRNOT00000075779
|
Olfr883
|
olfactory receptor 883 |
| chr14_-_21127868 | 4.73 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr1_-_256013495 | 4.72 |
ENSRNOT00000023342
|
Ide
|
insulin degrading enzyme |
| chr1_-_100671074 | 4.66 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chr6_+_54488038 | 4.61 |
ENSRNOT00000084661
ENSRNOT00000005637 |
Snx13
|
sorting nexin 13 |
| chr10_-_16046033 | 4.60 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr11_-_86328469 | 4.57 |
ENSRNOT00000071493
|
Ufd1l
|
ubiquitin fusion degradation 1 like (yeast) |
| chr8_-_43055175 | 4.50 |
ENSRNOT00000074413
|
LOC103693054
|
olfactory receptor 8G1-like |
| chr18_-_59975192 | 4.46 |
ENSRNOT00000024328
|
Fech
|
ferrochelatase |
| chr7_-_26984400 | 4.44 |
ENSRNOT00000013613
|
Txnrd1
|
thioredoxin reductase 1 |
| chr1_+_198690794 | 4.43 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr3_-_23020441 | 4.42 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr10_+_110346453 | 4.38 |
ENSRNOT00000054928
|
Tex19.1
|
testis expressed 19.1 |
| chrX_+_62754634 | 4.36 |
ENSRNOT00000016669
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr7_+_2479311 | 4.35 |
ENSRNOT00000003787
|
Ptges3
|
prostaglandin E synthase 3 |
| chr8_+_43566788 | 4.35 |
ENSRNOT00000087263
|
Olr1316
|
olfactory receptor 1316 |
| chr3_-_72289310 | 4.34 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr15_-_41316203 | 4.32 |
ENSRNOT00000065261
|
Tnfrsf19
|
TNF receptor superfamily member 19 |
| chr7_+_11152038 | 4.30 |
ENSRNOT00000006168
|
Nfic
|
nuclear factor I/C |
| chr1_+_105094411 | 4.27 |
ENSRNOT00000036258
|
Htatip2
|
HIV-1 Tat interactive protein 2 |
| chr4_+_71376535 | 4.26 |
ENSRNOT00000021832
|
Sval1
|
seminal vesicle antigen-like 1 |
| chr20_-_33322966 | 4.25 |
ENSRNOT00000000459
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr15_+_35843533 | 4.11 |
ENSRNOT00000087987
|
AC114070.2
|
|
| chr3_+_58164931 | 4.10 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr1_+_100297152 | 4.09 |
ENSRNOT00000026100
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr1_-_170318935 | 4.07 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr1_+_25174456 | 4.06 |
ENSRNOT00000092830
|
Clvs2
|
clavesin 2 |
| chr2_+_227455722 | 4.02 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr15_-_52317219 | 4.02 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr3_+_175548174 | 4.02 |
ENSRNOT00000091941
|
Adrm1
|
adhesion regulating molecule 1 |
| chr5_+_90338795 | 3.99 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr19_+_52566607 | 3.96 |
ENSRNOT00000022432
|
Usp10
|
ubiquitin specific peptidase 10 |
| chr16_-_84939323 | 3.94 |
ENSRNOT00000022742
|
Myo16
|
myosin XVI |
| chr8_+_80965255 | 3.92 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr1_+_238843338 | 3.87 |
ENSRNOT00000024583
|
Zfand5
|
zinc finger AN1-type containing 5 |
| chr3_+_111597102 | 3.85 |
ENSRNOT00000081462
|
Tyro3
|
TYRO3 protein tyrosine kinase |
| chr9_-_113328428 | 3.85 |
ENSRNOT00000086340
|
Vapa
|
VAMP associated protein A |
| chr8_+_116324040 | 3.84 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr3_+_103747654 | 3.80 |
ENSRNOT00000006887
|
Nop10
|
NOP10 ribonucleoprotein |
| chr3_+_58084606 | 3.79 |
ENSRNOT00000084797
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr8_+_43590248 | 3.78 |
ENSRNOT00000088053
|
Olr1318
|
olfactory receptor 1318 |
| chr20_-_33323367 | 3.76 |
ENSRNOT00000080444
|
Ros1
|
ROS proto-oncogene 1 , receptor tyrosine kinase |
| chr3_+_76790186 | 3.72 |
ENSRNOT00000084374
|
Olr635
|
olfactory receptor 635 |
| chr1_+_69615603 | 3.69 |
ENSRNOT00000084806
|
Zfp418
|
zinc finger protein 418 |
| chr8_+_40410604 | 3.64 |
ENSRNOT00000049392
|
Olr1202
|
olfactory receptor 1202 |
| chr10_+_94445877 | 3.64 |
ENSRNOT00000013997
|
Psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr3_+_2648885 | 3.60 |
ENSRNOT00000020339
|
Abca2
|
ATP binding cassette subfamily A member 2 |
| chr19_-_37250919 | 3.60 |
ENSRNOT00000021015
|
Exoc3l1
|
exocyst complex component 3-like 1 |
| chr4_+_1363724 | 3.56 |
ENSRNOT00000072782
|
LOC100910863
|
olfactory receptor 143-like |
| chr20_-_556982 | 3.55 |
ENSRNOT00000047478
|
Olr1683
|
olfactory receptor 1683 |
| chr10_+_46906115 | 3.48 |
ENSRNOT00000034006
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr19_-_26053762 | 3.47 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr20_-_3818045 | 3.46 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr8_-_62987182 | 3.45 |
ENSRNOT00000070885
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr16_-_755990 | 3.44 |
ENSRNOT00000013501
|
Polr3a
|
RNA polymerase III subunit A |
| chr8_+_40524733 | 3.37 |
ENSRNOT00000076626
ENSRNOT00000074528 |
Olr1206
|
olfactory receptor 1206 |
| chr7_-_63728988 | 3.37 |
ENSRNOT00000009645
|
Xpot
|
exportin for tRNA |
| chr6_-_86822094 | 3.36 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr3_+_1413671 | 3.33 |
ENSRNOT00000007675
|
Il1f10
|
interleukin 1 family member 10 |
| chr2_+_188748359 | 3.31 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr1_-_175676699 | 3.31 |
ENSRNOT00000030474
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr2_-_61414038 | 3.30 |
ENSRNOT00000051910
ENSRNOT00000088202 |
Tars
|
threonyl-tRNA synthetase |
| chr8_+_41954140 | 3.25 |
ENSRNOT00000049709
|
Olr1223
|
olfactory receptor 1223 |
| chr6_+_136330383 | 3.22 |
ENSRNOT00000093106
ENSRNOT00000092774 ENSRNOT00000065281 ENSRNOT00000015935 |
Klc1
|
kinesin light chain 1 |
| chr8_+_119228612 | 3.22 |
ENSRNOT00000078439
ENSRNOT00000043737 |
Lrrc2
|
leucine rich repeat containing 2 |
| chr1_-_78997869 | 3.22 |
ENSRNOT00000023490
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr11_-_83944763 | 3.20 |
ENSRNOT00000002358
|
Psmd2
|
proteasome 26S subunit, non-ATPase 2 |
| chr8_+_43644822 | 3.18 |
ENSRNOT00000085286
|
Olr1321
|
olfactory receptor 1321 |
| chr8_+_43675950 | 3.18 |
ENSRNOT00000079430
|
Olr1323
|
olfactory receptor 1323 |
| chr3_-_2490392 | 3.12 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr20_-_35342 | 3.11 |
ENSRNOT00000048277
|
LOC100910263
|
olfactory receptor 2B11-like |
| chr11_-_84037938 | 3.10 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr4_+_167351697 | 3.07 |
ENSRNOT00000007659
ENSRNOT00000046622 |
Grpcb
|
glutamine/glutamic acid-rich protein B |
| chr9_-_82154266 | 3.05 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr4_+_113866804 | 3.03 |
ENSRNOT00000081196
|
Loxl3
|
lysyl oxidase-like 3 |
| chr20_+_386421 | 3.02 |
ENSRNOT00000044420
|
Olr1670
|
olfactory receptor 1670 |
| chr4_-_55740627 | 3.00 |
ENSRNOT00000010658
|
Pax4
|
paired box 4 |
| chr16_+_21050243 | 2.99 |
ENSRNOT00000064308
|
Ncan
|
neurocan |
| chr3_-_158985814 | 2.97 |
ENSRNOT00000071070
|
LOC100909423
|
olfactory receptor 150-like |
| chr15_+_35761388 | 2.96 |
ENSRNOT00000086118
|
Olr1286
|
olfactory receptor 1286 |
| chr4_-_9881484 | 2.95 |
ENSRNOT00000016450
|
Psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr10_-_86393141 | 2.92 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr4_+_24222500 | 2.89 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr17_+_4165041 | 2.89 |
ENSRNOT00000061427
|
Tpbpa
|
trophoblast specific protein alpha |
| chr8_+_40383918 | 2.87 |
ENSRNOT00000071422
|
Olr1201
|
olfactory receptor 1201 |
| chr12_-_50404550 | 2.83 |
ENSRNOT00000073072
|
Crybb1
|
crystallin, beta B1 |
| chr5_+_120340646 | 2.80 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr10_+_95706685 | 2.73 |
ENSRNOT00000004283
|
Psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr1_-_225035687 | 2.68 |
ENSRNOT00000026539
|
Gng3
|
G protein subunit gamma 3 |
| chr2_+_46186105 | 2.62 |
ENSRNOT00000071256
|
LOC100910378
|
olfactory receptor 145-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2_Bach1_Mafk
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.1 | 48.2 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 11.9 | 59.3 | GO:0009115 | xanthine catabolic process(GO:0009115) |
| 9.4 | 28.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 7.7 | 23.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 6.2 | 24.7 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 5.8 | 29.0 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 5.7 | 22.8 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 5.4 | 21.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 5.0 | 15.0 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 4.5 | 49.7 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 4.5 | 58.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 4.3 | 12.8 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 4.0 | 12.1 | GO:2000158 | flavin-containing compound metabolic process(GO:0042726) positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 3.2 | 12.9 | GO:1904975 | response to bleomycin(GO:1904975) |
| 3.2 | 9.6 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 2.7 | 27.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 2.6 | 10.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 2.5 | 7.5 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 2.4 | 26.0 | GO:0015747 | urate transport(GO:0015747) |
| 2.3 | 9.2 | GO:0038086 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 2.3 | 6.9 | GO:0072162 | regulation of metanephros size(GO:0035566) metanephric mesenchymal cell differentiation(GO:0072162) |
| 2.2 | 6.6 | GO:1903233 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 2.2 | 36.8 | GO:0019532 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 2.1 | 19.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 2.1 | 8.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 2.0 | 6.0 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 2.0 | 8.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 1.9 | 9.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.9 | 57.4 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 1.8 | 7.1 | GO:1990375 | baculum development(GO:1990375) |
| 1.8 | 17.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 1.7 | 6.8 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.7 | 5.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 1.6 | 17.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.6 | 4.7 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 1.6 | 11.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.5 | 3.0 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 1.5 | 5.9 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 1.4 | 10.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.4 | 15.5 | GO:0051132 | NK T cell activation(GO:0051132) |
| 1.4 | 8.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 1.4 | 2.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 1.4 | 4.1 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 1.3 | 4.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 1.3 | 3.8 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.3 | 3.8 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 1.2 | 3.6 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.2 | 14.1 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.1 | 3.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 1.1 | 4.5 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 1.1 | 4.4 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 1.1 | 3.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 1.1 | 6.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 1.0 | 4.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.0 | 39.7 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 1.0 | 8.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.9 | 3.5 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.8 | 15.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.8 | 3.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.8 | 2.3 | GO:0015820 | leucine transport(GO:0015820) |
| 0.8 | 3.8 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.7 | 1.5 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.7 | 12.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.7 | 2.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.7 | 2.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.7 | 2.2 | GO:0042628 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.7 | 7.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.7 | 2.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.7 | 2.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.6 | 3.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.6 | 5.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.6 | 4.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.6 | 4.4 | GO:0060720 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.6 | 8.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.6 | 22.0 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.6 | 7.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 1.7 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.6 | 3.9 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.5 | 2.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.5 | 103.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.5 | 2.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.5 | 7.7 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 1.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic genitalia morphogenesis(GO:0030538) |
| 0.5 | 4.4 | GO:1904116 | response to vasopressin(GO:1904116) |
| 0.5 | 1.9 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.4 | 6.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 2.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.4 | 8.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.4 | 3.9 | GO:0060068 | vagina development(GO:0060068) |
| 0.4 | 3.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 43.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 20.3 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.4 | 1.8 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 2.5 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.4 | 4.4 | GO:0060430 | lung saccule development(GO:0060430) |
| 0.4 | 8.7 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 | 5.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 4.3 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.3 | 9.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.3 | 3.8 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.3 | 1.5 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.3 | 7.9 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.3 | 4.7 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.3 | 9.7 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 5.0 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.3 | 1.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.3 | 3.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 3.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 2.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 7.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 0.9 | GO:1905150 | regulation of membrane depolarization during action potential(GO:0098902) regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 2.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 3.9 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 8.2 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.2 | 0.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 3.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 3.9 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.4 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 1.0 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 2.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 38.6 | GO:0006914 | autophagy(GO:0006914) |
| 0.1 | 1.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 1.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 4.1 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 7.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 2.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 3.9 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 2.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 110.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 5.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 16.0 | GO:0001889 | liver development(GO:0001889) |
| 0.1 | 5.0 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 3.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 11.0 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 4.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 1.2 | GO:0071577 | cellular zinc ion homeostasis(GO:0006882) zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 4.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 23.0 | GO:0030163 | protein catabolic process(GO:0030163) |
| 0.0 | 0.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.5 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 3.0 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 7.2 | GO:0042692 | muscle cell differentiation(GO:0042692) |
| 0.0 | 2.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.4 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.8 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 4.0 | 12.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 3.4 | 58.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 3.0 | 48.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 2.6 | 12.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 2.5 | 22.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 2.3 | 36.6 | GO:0031045 | dense core granule(GO:0031045) |
| 2.2 | 11.0 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 2.2 | 6.6 | GO:0090534 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 1.5 | 30.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 1.5 | 1.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.5 | 7.3 | GO:0089701 | U2AF(GO:0089701) |
| 1.4 | 38.8 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 1.3 | 4.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.3 | 41.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 1.3 | 19.1 | GO:0044754 | autolysosome(GO:0044754) |
| 1.2 | 4.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.0 | 18.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.9 | 17.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 9.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.8 | 3.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.7 | 9.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.7 | 13.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.7 | 7.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.7 | 5.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.7 | 14.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.7 | 7.5 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.5 | 27.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.5 | 15.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.5 | 8.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.4 | 10.1 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 9.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 3.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 4.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 4.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 11.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 6.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.2 | 4.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 3.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 9.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 11.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 28.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 6.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 11.4 | GO:0044438 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.2 | 1.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 10.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.9 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 3.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 4.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 28.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 8.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 7.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 9.1 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 6.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 68.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 11.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 10.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 3.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 37.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 4.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 111.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 2.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 37.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 7.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 4.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.5 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.6 | 49.7 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 12.1 | 60.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 11.9 | 59.3 | GO:0004854 | aldehyde oxidase activity(GO:0004031) xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 9.4 | 28.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 8.3 | 58.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 6.4 | 25.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 5.7 | 22.8 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 4.5 | 27.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 4.5 | 40.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 4.3 | 12.8 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 4.0 | 12.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 3.7 | 15.0 | GO:0051870 | methotrexate binding(GO:0051870) |
| 3.2 | 16.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 3.1 | 21.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 3.1 | 15.5 | GO:0005534 | galactose binding(GO:0005534) |
| 3.1 | 55.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 3.1 | 9.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 2.9 | 8.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 2.6 | 26.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 2.5 | 36.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 2.0 | 5.9 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 1.5 | 12.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.5 | 8.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.5 | 4.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.4 | 7.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.4 | 39.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 1.3 | 10.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.3 | 3.8 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.2 | 17.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 1.2 | 24.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 1.2 | 3.5 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 1.1 | 4.5 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 1.1 | 3.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 1.0 | 8.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 1.0 | 8.7 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.0 | 3.9 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.9 | 24.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 6.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.9 | 4.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.9 | 3.5 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.8 | 5.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.8 | 3.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.8 | 43.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.7 | 23.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.7 | 4.4 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.7 | 7.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.7 | 18.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.7 | 7.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.7 | 19.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.6 | 24.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.6 | 6.4 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.6 | 7.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.6 | 3.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.6 | 4.1 | GO:0071532 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 0.6 | 100.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.6 | 4.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 14.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.5 | 4.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.5 | 7.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.5 | 2.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 5.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 4.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 6.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 3.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 3.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.4 | 1.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.4 | 10.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 11.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 2.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.3 | 4.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 4.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.3 | 7.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 1.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 21.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.3 | 2.7 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 3.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 6.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 13.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 0.9 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.2 | 2.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 5.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 18.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 3.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 4.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 3.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 2.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 4.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 2.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 64.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 4.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.5 | GO:0045295 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.1 | 2.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 9.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 15.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 13.8 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 1.9 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 3.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 13.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 9.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 9.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 2.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 4.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 2.2 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 4.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 46.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 4.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 6.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.0 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 2.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 7.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 3.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 5.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 7.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.1 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 12.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.8 | 62.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.8 | 40.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.5 | 9.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.5 | 20.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.5 | 19.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.4 | 5.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 23.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 12.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.3 | 9.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.3 | 13.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 24.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.3 | 3.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 8.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.3 | 12.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.3 | 10.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 3.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 4.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 4.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 13.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 46.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 2.4 | 26.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.7 | 90.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 1.6 | 19.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 1.5 | 58.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.1 | 12.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.9 | 24.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.8 | 24.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 28.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.5 | 7.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 9.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 12.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.5 | 23.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.5 | 9.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.5 | 8.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 54.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.4 | 54.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.4 | 9.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.4 | 5.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.3 | 6.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 10.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.3 | 33.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 25.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.3 | 4.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 3.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 7.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 4.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 3.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 4.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 7.0 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.1 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 7.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 4.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |