Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
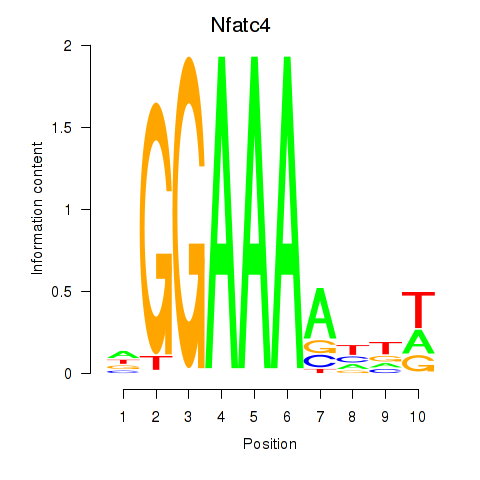
Results for Nfatc4
Z-value: 0.17
Transcription factors associated with Nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc4
|
ENSRNOG00000020482 | nuclear factor of activated T-cells 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc4 | rn6_v1_chr15_+_34493138_34493163 | 0.24 | 1.3e-05 | Click! |
Activity profile of Nfatc4 motif
Sorted Z-values of Nfatc4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_32019205 | 3.24 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr2_+_149315189 | 2.93 |
ENSRNOT00000082464
|
Med12l
|
mediator complex subunit 12-like |
| chr1_-_270472866 | 2.92 |
ENSRNOT00000015979
|
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr3_-_52510507 | 2.71 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr4_-_159697207 | 1.56 |
ENSRNOT00000086440
|
Ccnd2
|
cyclin D2 |
| chr10_-_38969501 | 1.41 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr2_+_11658568 | 1.27 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_184263564 | 1.06 |
ENSRNOT00000015279
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr3_+_8873933 | 0.99 |
ENSRNOT00000030996
|
Nup188
|
nucleoporin 188 |
| chr2_-_123851854 | 0.91 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr5_-_28130803 | 0.84 |
ENSRNOT00000093186
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr15_+_24479761 | 0.73 |
ENSRNOT00000016449
|
Ktn1
|
kinectin 1 |
| chr8_-_36374673 | 0.71 |
ENSRNOT00000013763
|
Dcps
|
decapping enzyme, scavenger |
| chr10_-_32471454 | 0.62 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr3_-_120157866 | 0.62 |
ENSRNOT00000020367
|
LOC100909998
|
zinc finger protein 2-like |
| chr7_+_35773928 | 0.59 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr2_-_247446882 | 0.47 |
ENSRNOT00000021963
|
Bmpr1b
|
bone morphogenetic protein receptor type 1B |
| chr9_+_53013413 | 0.40 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr6_+_95826546 | 0.25 |
ENSRNOT00000043587
|
AABR07064810.1
|
|
| chr12_-_7186873 | 0.22 |
ENSRNOT00000050438
|
LOC103690996
|
uncharacterized LOC103690996 |
| chr1_+_230268991 | 0.11 |
ENSRNOT00000083727
|
Olr363
|
olfactory receptor 363 |
| chr17_-_14557792 | 0.06 |
ENSRNOT00000078166
|
LOC103690033
|
isoleucine--tRNA ligase, cytoplasmic-like |
| chr6_+_115170865 | 0.00 |
ENSRNOT00000005671
|
Tshr
|
thyroid stimulating hormone receptor |
| chr8_+_41365984 | 0.00 |
ENSRNOT00000074250
|
Olr1228
|
olfactory receptor 1228 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.5 | 1.4 | GO:2000424 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.4 | 1.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 1.3 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 1.0 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 0.9 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 2.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.6 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.6 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.5 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.8 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 3.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 2.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 1.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.7 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |