Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
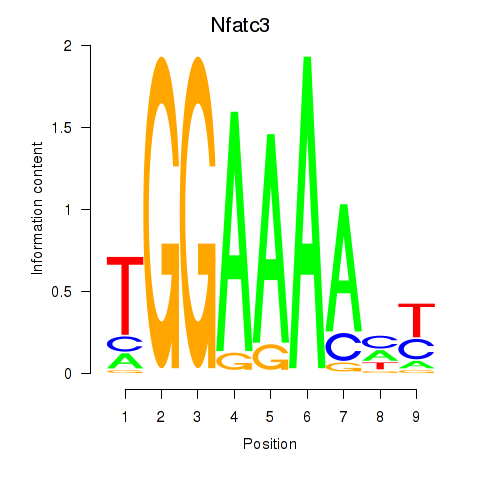
Results for Nfatc3
Z-value: 0.68
Transcription factors associated with Nfatc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc3
|
ENSRNOG00000054264 | nuclear factor of activated T-cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc3 | rn6_v1_chr19_+_38039564_38039564 | -0.03 | 6.4e-01 | Click! |
Activity profile of Nfatc3 motif
Sorted Z-values of Nfatc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_138483612 | 34.80 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr2_-_182035032 | 26.91 |
ENSRNOT00000009813
|
Fgb
|
fibrinogen beta chain |
| chr2_+_235264219 | 22.89 |
ENSRNOT00000086245
|
Cfi
|
complement factor I |
| chr13_+_91080341 | 20.87 |
ENSRNOT00000000058
|
Crp
|
C-reactive protein |
| chr2_+_198655437 | 18.30 |
ENSRNOT00000028781
|
Hfe2
|
hemochromatosis type 2 (juvenile) |
| chr2_+_243502073 | 13.87 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr1_-_166037424 | 13.35 |
ENSRNOT00000026115
|
P2ry2
|
purinergic receptor P2Y2 |
| chr16_-_7007051 | 13.12 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr16_-_7007287 | 12.90 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr9_+_14529218 | 11.69 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr2_-_105089659 | 11.18 |
ENSRNOT00000043381
|
Cpb1
|
carboxypeptidase B1 |
| chr1_+_279798187 | 10.53 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr10_-_41408518 | 9.83 |
ENSRNOT00000018967
|
Nmur2
|
neuromedin U receptor 2 |
| chr12_+_24989298 | 9.64 |
ENSRNOT00000032780
|
Eln
|
elastin |
| chr17_-_35958077 | 9.43 |
ENSRNOT00000038532
|
Agtr1a
|
angiotensin II receptor, type 1a |
| chr18_+_53915807 | 9.40 |
ENSRNOT00000026543
|
Adamts19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19 |
| chr3_-_71845232 | 9.19 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr1_+_151918571 | 9.04 |
ENSRNOT00000022342
|
Ctsc
|
cathepsin C |
| chr1_+_166433109 | 8.90 |
ENSRNOT00000026428
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr18_-_59819113 | 8.83 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr1_+_189328246 | 8.45 |
ENSRNOT00000084260
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr13_-_98478327 | 8.21 |
ENSRNOT00000030135
|
Coq8a
|
coenzyme Q8A |
| chr4_+_49056010 | 7.71 |
ENSRNOT00000038566
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr3_-_48535909 | 7.60 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr15_+_10120206 | 7.48 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr18_-_77322690 | 7.33 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr15_-_14510828 | 7.18 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr19_-_1074333 | 7.07 |
ENSRNOT00000017983
ENSRNOT00000086995 |
Cdh5
|
cadherin 5 |
| chr9_+_98924134 | 7.06 |
ENSRNOT00000027597
|
Twist2
|
twist family bHLH transcription factor 2 |
| chr16_+_10417185 | 6.99 |
ENSRNOT00000082186
|
Anxa8
|
annexin A8 |
| chr11_+_7265828 | 6.88 |
ENSRNOT00000084765
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr20_-_49486550 | 6.63 |
ENSRNOT00000048270
ENSRNOT00000076541 |
Prdm1
|
PR/SET domain 1 |
| chr3_-_110324084 | 6.63 |
ENSRNOT00000010381
|
Bmf
|
Bcl2 modifying factor |
| chr20_+_26988774 | 6.58 |
ENSRNOT00000090083
|
Mypn
|
myopalladin |
| chr8_-_114449956 | 6.57 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr11_+_61609370 | 6.42 |
ENSRNOT00000088880
ENSRNOT00000082533 |
Gramd1c
|
GRAM domain containing 1C |
| chr3_+_14482388 | 6.32 |
ENSRNOT00000025857
|
Gsn
|
gelsolin |
| chr3_-_48536235 | 6.17 |
ENSRNOT00000083536
|
Fap
|
fibroblast activation protein, alpha |
| chr16_+_59077574 | 6.12 |
ENSRNOT00000090477
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr11_-_38274217 | 6.02 |
ENSRNOT00000002206
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr6_+_2216623 | 5.88 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr12_-_41307285 | 5.87 |
ENSRNOT00000075317
ENSRNOT00000048782 ENSRNOT00000075234 |
Oas1b
|
2-5 oligoadenylate synthetase 1B |
| chr11_-_60679555 | 5.84 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr2_-_197814808 | 5.82 |
ENSRNOT00000074156
|
Adamtsl4
|
ADAMTS-like 4 |
| chr9_-_100306829 | 5.78 |
ENSRNOT00000038563
|
RGD1563692
|
similar to hypothetical protein FLJ22671 |
| chr8_-_18508971 | 5.75 |
ENSRNOT00000061339
|
Muc16
|
mucin 16, cell surface associated |
| chr13_+_91481461 | 5.60 |
ENSRNOT00000012103
|
Olr1576
|
olfactory receptor 1576 |
| chr17_-_81187739 | 5.34 |
ENSRNOT00000063911
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr10_-_32471454 | 5.27 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr4_-_176922424 | 5.01 |
ENSRNOT00000055540
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr16_-_47537476 | 4.94 |
ENSRNOT00000050279
|
Cldn24
|
claudin 24 |
| chr17_-_51912496 | 4.77 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr13_+_89586283 | 4.63 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr2_+_195617021 | 4.47 |
ENSRNOT00000067042
ENSRNOT00000036656 |
Rorc
Lingo4
|
RAR-related orphan receptor C leucine rich repeat and Ig domain containing 4 |
| chr10_+_83655460 | 4.43 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr9_+_95221474 | 4.20 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr14_-_72025137 | 4.19 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr16_-_31301880 | 4.18 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr5_+_173256834 | 4.12 |
ENSRNOT00000089936
|
Ccnl2
|
cyclin L2 |
| chr10_-_74769637 | 4.10 |
ENSRNOT00000008889
|
LOC102555183
|
zinc finger protein OZF-like |
| chr5_-_164747083 | 4.09 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr10_+_90930010 | 4.08 |
ENSRNOT00000003765
|
Higd1b
|
HIG1 hypoxia inducible domain family, member 1B |
| chr4_+_180291389 | 4.07 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr11_-_62128044 | 4.06 |
ENSRNOT00000093141
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr5_+_152533349 | 4.00 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr1_+_177569618 | 3.95 |
ENSRNOT00000090042
|
Tead1
|
TEA domain transcription factor 1 |
| chr2_-_251532312 | 3.86 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr14_-_18591394 | 3.83 |
ENSRNOT00000003716
|
Ereg
|
epiregulin |
| chr6_+_64649194 | 3.78 |
ENSRNOT00000039776
|
AABR07064102.1
|
|
| chr7_-_36499784 | 3.77 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr12_-_51341663 | 3.67 |
ENSRNOT00000066892
ENSRNOT00000052202 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr5_+_173256637 | 3.49 |
ENSRNOT00000025531
|
Ccnl2
|
cyclin L2 |
| chr14_-_112946204 | 3.46 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr1_+_163398891 | 3.40 |
ENSRNOT00000020513
|
Gucy2e
|
guanylate cyclase 2E |
| chr10_-_103687425 | 3.36 |
ENSRNOT00000039284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr7_-_117267402 | 3.32 |
ENSRNOT00000088945
|
Plec
|
plectin |
| chr1_+_171797516 | 3.31 |
ENSRNOT00000088110
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr15_-_34469350 | 3.24 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr14_+_7618022 | 3.24 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr1_+_41192824 | 3.24 |
ENSRNOT00000082133
|
Esr1
|
estrogen receptor 1 |
| chr7_-_117267803 | 3.23 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr14_+_5928737 | 3.21 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr15_+_32763067 | 3.21 |
ENSRNOT00000011998
|
AABR07017902.1
|
|
| chr11_-_65845418 | 3.05 |
ENSRNOT00000091057
|
Fstl1
|
follistatin-like 1 |
| chr12_-_30083824 | 3.01 |
ENSRNOT00000084968
|
Tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr12_+_13102019 | 2.95 |
ENSRNOT00000092628
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr4_+_99399148 | 2.94 |
ENSRNOT00000088018
|
Rnf103
|
ring finger protein 103 |
| chr4_+_22084954 | 2.91 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr3_-_151486693 | 2.89 |
ENSRNOT00000073736
ENSRNOT00000071099 |
Gdf5
|
growth differentiation factor 5 |
| chr17_-_22143324 | 2.73 |
ENSRNOT00000019361
|
Edn1
|
endothelin 1 |
| chr2_-_45077219 | 2.71 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr1_-_266914093 | 2.64 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr4_-_2381271 | 2.48 |
ENSRNOT00000072287
|
Mnx1
|
motor neuron and pancreas homeobox 1 |
| chr3_+_73390268 | 2.46 |
ENSRNOT00000052240
|
Olr475
|
olfactory receptor 475 |
| chr3_-_69637377 | 2.38 |
ENSRNOT00000074765
|
AABR07052692.1
|
|
| chr18_+_76770012 | 2.37 |
ENSRNOT00000089389
|
Pqlc1
|
PQ loop repeat containing 1 |
| chr16_+_54332660 | 2.37 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr7_-_132343169 | 2.36 |
ENSRNOT00000021064
|
Abcd2
|
ATP binding cassette subfamily D member 2 |
| chr6_+_135890931 | 2.36 |
ENSRNOT00000042973
|
Tnfaip2
|
TNF alpha induced protein 2 |
| chr11_+_84094520 | 2.33 |
ENSRNOT00000046642
|
Rps15al2
|
ribosomal protein S15A-like 2 |
| chr2_-_174413038 | 2.30 |
ENSRNOT00000089229
ENSRNOT00000037115 |
Golim4
|
golgi integral membrane protein 4 |
| chr7_-_54778848 | 2.28 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr13_-_83202864 | 2.27 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr15_-_34444244 | 2.24 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr7_-_29701586 | 2.23 |
ENSRNOT00000009084
ENSRNOT00000089269 |
Ano4
|
anoctamin 4 |
| chr9_-_78693028 | 2.20 |
ENSRNOT00000036359
|
Abca12
|
ATP binding cassette subfamily A member 12 |
| chr3_+_113251778 | 2.19 |
ENSRNOT00000083005
|
Map1a
|
microtubule-associated protein 1A |
| chr16_-_6404578 | 2.12 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr16_-_47535358 | 2.06 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr1_+_37507276 | 2.05 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chr6_+_95816749 | 2.02 |
ENSRNOT00000008880
|
Six6
|
SIX homeobox 6 |
| chrX_-_100207974 | 1.99 |
ENSRNOT00000091859
|
AABR07040430.1
|
|
| chr1_+_164706276 | 1.94 |
ENSRNOT00000024311
|
Olr36
|
olfactory receptor 36 |
| chr7_+_27438351 | 1.94 |
ENSRNOT00000077976
|
AABR07056503.1
|
|
| chr2_+_212435728 | 1.89 |
ENSRNOT00000046960
|
AABR07012868.1
|
|
| chrX_+_83678339 | 1.85 |
ENSRNOT00000005997
|
Apool
|
apolipoprotein O-like |
| chr8_+_42322813 | 1.75 |
ENSRNOT00000040431
|
Olr1241
|
olfactory receptor 1241 |
| chr10_-_87891285 | 1.73 |
ENSRNOT00000090052
|
Rn50_10_0877.5
|
|
| chr6_-_128894761 | 1.70 |
ENSRNOT00000029346
|
Tcl1a
|
T-cell leukemia/lymphoma 1A |
| chr5_+_107233230 | 1.68 |
ENSRNOT00000029976
|
Ifne
|
interferon, epsilon |
| chr6_+_137063611 | 1.65 |
ENSRNOT00000017504
|
LOC691485
|
hypothetical protein LOC691485 |
| chr10_+_3655038 | 1.65 |
ENSRNOT00000045047
|
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr6_-_1454480 | 1.64 |
ENSRNOT00000072810
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr8_+_20780898 | 1.64 |
ENSRNOT00000052197
|
LOC690821
|
similar to olfactory receptor Olr1185 |
| chr13_+_107580629 | 1.63 |
ENSRNOT00000003557
|
LOC100361993
|
transmembrane protein 14C |
| chr1_+_230160365 | 1.62 |
ENSRNOT00000088190
|
Olr361
|
olfactory receptor 361 |
| chr3_-_176958880 | 1.60 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr8_+_116776494 | 1.59 |
ENSRNOT00000050702
|
Cdhr4
|
cadherin-related family member 4 |
| chr1_-_149778698 | 1.59 |
ENSRNOT00000089920
|
LOC100909935
|
olfactory receptor 6F1-like |
| chr3_-_93216495 | 1.59 |
ENSRNOT00000010580
|
Ehf
|
ets homologous factor |
| chr3_+_67849966 | 1.58 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr12_-_51965779 | 1.58 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr5_-_58078545 | 1.55 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr2_-_199563386 | 1.55 |
ENSRNOT00000075261
|
AABR07012616.1
|
|
| chr13_+_90116843 | 1.54 |
ENSRNOT00000006306
|
Cd48
|
Cd48 molecule |
| chr4_-_165774126 | 1.51 |
ENSRNOT00000007453
|
Tas2r107
|
taste receptor, type 2, member 107 |
| chr2_-_198458041 | 1.50 |
ENSRNOT00000082450
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr17_-_27665266 | 1.49 |
ENSRNOT00000060218
|
Rreb1
|
ras responsive element binding protein 1 |
| chr4_-_56114254 | 1.49 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr3_-_112061796 | 1.47 |
ENSRNOT00000056258
|
Pla2g4d
|
phospholipase A2 group IVD |
| chr1_-_230649938 | 1.46 |
ENSRNOT00000017615
|
Olr377
|
olfactory receptor 377 |
| chr8_+_61603991 | 1.45 |
ENSRNOT00000065156
|
AC135389.1
|
|
| chr16_-_10603850 | 1.42 |
ENSRNOT00000087737
|
Fam35a
|
family with sequence similarity 35, member A |
| chr1_-_2017488 | 1.39 |
ENSRNOT00000021905
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr3_+_114087287 | 1.39 |
ENSRNOT00000023017
|
B2m
|
beta-2 microglobulin |
| chr1_+_16910069 | 1.37 |
ENSRNOT00000020015
|
Aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr2_-_35229353 | 1.37 |
ENSRNOT00000085814
|
LOC100910041
|
olfactory receptor 146-like |
| chr3_-_103066392 | 1.37 |
ENSRNOT00000044181
|
Olr776
|
olfactory receptor 776 |
| chr10_+_85257876 | 1.36 |
ENSRNOT00000014752
|
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr5_+_152466331 | 1.34 |
ENSRNOT00000082841
|
LOC108351043
|
uncharacterized LOC108351043 |
| chr7_+_6215743 | 1.34 |
ENSRNOT00000050967
|
Olr1002
|
olfactory receptor 1002 |
| chr3_-_46942966 | 1.31 |
ENSRNOT00000087439
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr2_+_206902962 | 1.29 |
ENSRNOT00000040909
|
RGD1564469
|
similar to Acidic ribosomal phosphoprotein P0 |
| chr15_-_31770690 | 1.24 |
ENSRNOT00000082666
|
AABR07017841.2
|
|
| chr1_+_229889771 | 1.16 |
ENSRNOT00000085941
|
Olr348
|
olfactory receptor 348 |
| chr1_+_267359830 | 1.15 |
ENSRNOT00000015495
|
Slk
|
STE20-like kinase |
| chr3_-_76279104 | 1.14 |
ENSRNOT00000007796
|
Olr608
|
olfactory receptor 608 |
| chr3_+_21016266 | 1.13 |
ENSRNOT00000010473
|
Olr420
|
olfactory receptor 420 |
| chr3_+_61597382 | 1.12 |
ENSRNOT00000068636
|
Hoxd12
|
homeo box D12 |
| chr1_+_196095214 | 1.11 |
ENSRNOT00000080741
|
LOC691716
|
similar to ribosomal protein S15a |
| chrX_-_115073890 | 1.09 |
ENSRNOT00000006638
|
Capn6
|
calpain 6 |
| chr12_+_4310334 | 1.09 |
ENSRNOT00000041244
|
Vom2r60
|
vomeronasal 2 receptor, 60 |
| chr13_-_89606326 | 1.08 |
ENSRNOT00000029179
|
Fcer1g
|
Fc fragment of IgE receptor Ig |
| chr14_-_44225713 | 1.08 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr2_+_188583664 | 1.08 |
ENSRNOT00000045477
|
Dpm3
|
dolichyl-phosphate mannosyltransferase subunit 3 |
| chr4_+_99168248 | 1.07 |
ENSRNOT00000058186
|
LOC102552663
|
uncharacterized LOC102552663 |
| chr3_-_47025128 | 1.07 |
ENSRNOT00000011682
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_-_71080499 | 1.04 |
ENSRNOT00000074623
|
RGD1562066
|
similar to odorant receptor ORZ6 |
| chrX_-_137015266 | 1.03 |
ENSRNOT00000040369
|
Olr1767
|
olfactory receptor 1767 |
| chr17_+_60287203 | 1.02 |
ENSRNOT00000025585
|
Armc4
|
armadillo repeat containing 4 |
| chr5_-_147784311 | 1.01 |
ENSRNOT00000074172
|
Fam167b
|
family with sequence similarity 167, member B |
| chr4_+_96562725 | 1.00 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr5_-_9429859 | 0.97 |
ENSRNOT00000009462
|
Adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr3_-_74815509 | 0.97 |
ENSRNOT00000045173
|
Olr499
|
olfactory receptor 499 |
| chr6_+_69971227 | 0.97 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr1_+_229755649 | 0.95 |
ENSRNOT00000046384
|
Olr340
|
olfactory receptor 340 |
| chr4_-_125808281 | 0.94 |
ENSRNOT00000037848
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_-_172932507 | 0.94 |
ENSRNOT00000048478
|
RGD1564933
|
similar to olfactory receptor Olr271 |
| chr12_+_3969493 | 0.93 |
ENSRNOT00000042692
|
Vom2r59
|
vomeronasal 2 receptor, 59 |
| chr17_-_81558565 | 0.92 |
ENSRNOT00000048715
|
AABR07028691.1
|
|
| chr11_-_45688560 | 0.92 |
ENSRNOT00000081508
|
Olr1536
|
olfactory receptor 1536 |
| chr1_+_213583606 | 0.92 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chr2_+_150106292 | 0.90 |
ENSRNOT00000073078
|
LOC100910567
|
arylacetamide deacetylase-like 2-like |
| chr1_+_174702373 | 0.89 |
ENSRNOT00000013733
|
Zfp143
|
zinc finger protein 143 |
| chr13_+_27238767 | 0.88 |
ENSRNOT00000003495
|
Serpinb11
|
serpin family B member 11 |
| chr8_-_43304560 | 0.87 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr1_-_230115342 | 0.87 |
ENSRNOT00000017444
|
Olr360
|
olfactory receptor 360 |
| chr2_-_250600517 | 0.86 |
ENSRNOT00000016872
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr1_-_6970040 | 0.85 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chr7_-_16432738 | 0.83 |
ENSRNOT00000049493
|
Olr932
|
olfactory receptor 932 |
| chr13_+_56513286 | 0.81 |
ENSRNOT00000015596
|
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr10_+_46511271 | 0.81 |
ENSRNOT00000080828
|
Rai1
|
retinoic acid induced 1 |
| chrX_-_138148967 | 0.81 |
ENSRNOT00000033968
|
Frmd7
|
FERM domain containing 7 |
| chr3_-_105298108 | 0.78 |
ENSRNOT00000010994
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr1_+_129255396 | 0.78 |
ENSRNOT00000040147
|
Fam169b
|
family with sequence similarity 169, member B |
| chr1_-_147272006 | 0.77 |
ENSRNOT00000086862
|
AABR07004539.1
|
|
| chr15_+_105640097 | 0.75 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chrX_-_105568343 | 0.74 |
ENSRNOT00000029807
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr13_-_91638661 | 0.74 |
ENSRNOT00000045429
|
Olr1579
|
olfactory receptor 1579 |
| chr1_-_6903486 | 0.73 |
ENSRNOT00000086574
|
Utrn
|
utrophin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 5.4 | 26.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 4.6 | 13.8 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 3.5 | 20.9 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 3.1 | 9.4 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 2.9 | 11.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 2.3 | 13.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 2.1 | 6.3 | GO:1903903 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 1.8 | 7.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.8 | 5.3 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.7 | 7.0 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 1.5 | 13.4 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 1.4 | 4.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.3 | 8.9 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 1.2 | 4.8 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 1.2 | 7.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.1 | 18.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 1.1 | 2.3 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 1.1 | 3.4 | GO:2000426 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.0 | 3.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 1.0 | 5.0 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 1.0 | 4.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 1.0 | 5.8 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.0 | 10.5 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.9 | 7.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.9 | 2.7 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.9 | 4.5 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.9 | 2.6 | GO:0002892 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) positive regulation of type IIa hypersensitivity(GO:0001798) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.8 | 6.8 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.8 | 6.6 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 0.8 | 26.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.8 | 3.2 | GO:1990375 | baculum development(GO:1990375) |
| 0.8 | 7.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.7 | 22.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 9.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.6 | 7.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.6 | 6.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.6 | 3.0 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.6 | 2.9 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.6 | 6.6 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.5 | 3.8 | GO:0045410 | oogenesis stage(GO:0022605) positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.5 | 8.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 4.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 1.4 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.4 | 2.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.4 | 2.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.4 | 1.6 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.4 | 1.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.4 | 6.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.4 | 4.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.3 | 6.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 2.4 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.3 | 1.6 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.3 | 25.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.3 | 0.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.7 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.2 | 9.0 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.2 | 2.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 0.7 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 0.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 2.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 1.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 0.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.4 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 8.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 2.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 0.7 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 0.5 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.7 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 1.9 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 4.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 6.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.2 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.1 | 2.2 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 8.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 9.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 2.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.0 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 0.2 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.1 | 1.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 3.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 5.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.8 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 2.2 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.6 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 1.8 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 5.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 2.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 1.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 5.8 | GO:0030198 | extracellular matrix organization(GO:0030198) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 5.2 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 31.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.6 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.6 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 1.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.4 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 3.2 | GO:0015711 | organic anion transport(GO:0015711) |
| 0.0 | 0.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 26.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 3.3 | 19.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 2.4 | 9.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 2.0 | 13.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 1.6 | 4.8 | GO:0043511 | inhibin complex(GO:0043511) |
| 1.3 | 4.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.3 | 5.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.1 | 6.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 5.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.7 | 8.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.7 | 9.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 11.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 1.6 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.5 | 2.7 | GO:0048237 | Weibel-Palade body(GO:0033093) rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 5.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 2.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 8.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 6.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 1.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 3.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 0.7 | GO:0042585 | germinal vesicle(GO:0042585) dendritic branch(GO:0044307) |
| 0.2 | 2.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 20.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 7.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 17.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 4.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 5.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 7.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 1.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 6.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.2 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 8.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 24.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 57.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 9.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.4 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 3.2 | 9.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 3.1 | 9.4 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 3.1 | 9.2 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 3.0 | 18.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 3.0 | 20.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 2.8 | 13.9 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 2.3 | 6.9 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 1.9 | 11.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.8 | 7.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 1.8 | 5.3 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 1.7 | 5.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 1.6 | 6.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.5 | 4.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.4 | 13.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.4 | 4.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 1.3 | 9.0 | GO:0031404 | chloride ion binding(GO:0031404) |
| 1.2 | 4.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.1 | 8.5 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 1.0 | 3.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.0 | 2.9 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.9 | 2.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.8 | 3.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.7 | 5.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.6 | 3.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.6 | 7.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.6 | 3.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.6 | 12.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 1.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.5 | 10.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 22.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.4 | 3.0 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.4 | 4.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.4 | 11.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 6.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.4 | 7.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.4 | 1.6 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.4 | 2.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.4 | 2.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.4 | 1.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.4 | 2.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.3 | 8.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 4.0 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 1.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 26.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 1.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 1.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.3 | 6.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 6.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 4.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 7.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 25.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.2 | 26.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 6.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 6.3 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.2 | 5.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.2 | 3.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 8.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 1.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 4.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 15.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 7.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 3.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.9 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 1.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 4.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.2 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 12.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 3.7 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.1 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 1.4 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 12.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 30.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 23.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.4 | 19.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.3 | 7.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 6.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 5.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 8.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 55.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 7.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 4.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 3.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 4.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 6.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 3.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 10.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 20.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.8 | 22.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.7 | 13.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.7 | 26.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.0 | 12.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.9 | 33.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 4.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 1.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 6.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 21.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.4 | 5.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 10.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 6.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 10.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 7.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 4.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 7.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 3.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 4.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 8.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 2.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 9.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 3.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 9.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 9.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 6.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 4.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 10.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.3 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 2.1 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 3.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 4.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |