Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
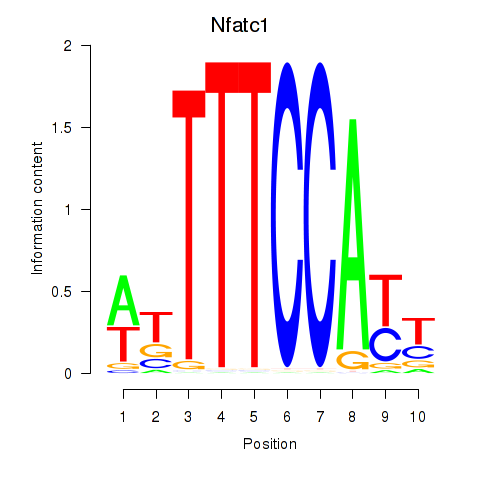
Results for Nfatc1
Z-value: 1.09
Transcription factors associated with Nfatc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc1
|
ENSRNOG00000017146 | nuclear factor of activated T-cells 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | rn6_v1_chr18_-_77322690_77322690 | 0.70 | 2.2e-49 | Click! |
Activity profile of Nfatc1 motif
Sorted Z-values of Nfatc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_80796670 | 40.56 |
ENSRNOT00000010539
|
Abra
|
actin-binding Rho activating protein |
| chr9_+_14529218 | 35.56 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr11_+_35568182 | 29.69 |
ENSRNOT00000002259
|
Kcnj15
|
potassium voltage-gated channel subfamily J member 15 |
| chr3_+_14482388 | 28.93 |
ENSRNOT00000025857
|
Gsn
|
gelsolin |
| chr7_+_38858062 | 27.18 |
ENSRNOT00000006234
|
Kera
|
keratocan |
| chr1_+_89008117 | 26.79 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr18_-_77322690 | 24.35 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chrX_-_31780425 | 23.76 |
ENSRNOT00000004693
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr8_-_114449956 | 22.61 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr2_-_257484587 | 22.21 |
ENSRNOT00000083344
|
Nexn
|
nexilin (F actin binding protein) |
| chr2_-_22744407 | 21.38 |
ENSRNOT00000073710
|
Cmya5
|
cardiomyopathy associated 5 |
| chr5_-_60191941 | 21.16 |
ENSRNOT00000033373
|
Pax5
|
paired box 5 |
| chr2_+_11658568 | 21.07 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr13_+_98615287 | 20.87 |
ENSRNOT00000004032
|
Itpkb
|
inositol-trisphosphate 3-kinase B |
| chr1_+_198655742 | 20.86 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr6_-_125723732 | 19.77 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr6_-_125723944 | 19.03 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr16_-_49574314 | 18.94 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr7_+_120153184 | 18.55 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr7_-_98098268 | 18.05 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr13_+_50164563 | 17.80 |
ENSRNOT00000029533
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr1_-_237910012 | 17.38 |
ENSRNOT00000023664
|
Anxa1
|
annexin A1 |
| chr1_+_18491384 | 17.07 |
ENSRNOT00000079138
ENSRNOT00000014917 |
Lama2
|
laminin subunit alpha 2 |
| chr7_-_81592206 | 16.81 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr16_-_75309176 | 16.55 |
ENSRNOT00000018427
|
Defb1
|
defensin beta 1 |
| chr4_+_98337367 | 16.27 |
ENSRNOT00000042165
|
AABR07060872.1
|
|
| chr4_-_176922424 | 16.17 |
ENSRNOT00000055540
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr2_-_251532312 | 16.12 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr3_-_71845232 | 15.97 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr14_-_22890958 | 14.89 |
ENSRNOT00000031916
|
LOC100911104
|
lymphocyte antigen 6B-like |
| chr4_+_153774486 | 14.88 |
ENSRNOT00000074096
|
Tuba8
|
tubulin, alpha 8 |
| chr6_+_10348308 | 14.43 |
ENSRNOT00000034991
|
Epas1
|
endothelial PAS domain protein 1 |
| chr4_+_180291389 | 14.43 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr13_-_82005741 | 14.36 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr16_+_6962722 | 14.07 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr13_-_82006005 | 13.61 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr13_+_90301006 | 13.55 |
ENSRNOT00000029315
|
Slamf6
|
SLAM family member 6 |
| chr2_+_190007216 | 13.32 |
ENSRNOT00000015612
|
S100a6
|
S100 calcium binding protein A6 |
| chr5_+_144308611 | 13.17 |
ENSRNOT00000014388
|
Col8a2
|
collagen type VIII alpha 2 chain |
| chr3_+_97349454 | 13.14 |
ENSRNOT00000089524
|
Dcdc5
|
doublecortin domain containing 5 |
| chr17_-_31813716 | 13.12 |
ENSRNOT00000074147
|
AABR07027451.1
|
|
| chr4_+_169147243 | 13.08 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr4_+_169161585 | 12.92 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr14_+_82867946 | 12.79 |
ENSRNOT00000075130
|
Spon2
|
spondin 2 |
| chr1_-_25839198 | 12.73 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr2_+_189423559 | 12.57 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr17_-_15511972 | 12.48 |
ENSRNOT00000074041
|
Aspnl1
|
asporin-like 1 |
| chr4_+_14109864 | 12.43 |
ENSRNOT00000076349
|
RGD1565355
|
similar to fatty acid translocase/CD36 |
| chr6_+_137063611 | 12.35 |
ENSRNOT00000017504
|
LOC691485
|
hypothetical protein LOC691485 |
| chr11_-_73678984 | 11.89 |
ENSRNOT00000002355
|
Fam43a
|
family with sequence similarity 43, member A |
| chr8_-_84632817 | 11.78 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr16_+_81616604 | 11.76 |
ENSRNOT00000026392
ENSRNOT00000057740 |
Adprhl1
Grtp1
|
ADP-ribosylhydrolase like 1 growth hormone regulated TBC protein 1 |
| chr1_-_266914093 | 11.68 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr10_+_23661013 | 11.58 |
ENSRNOT00000076664
|
Ebf1
|
early B-cell factor 1 |
| chr4_+_183656013 | 11.50 |
ENSRNOT00000055442
|
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr10_-_108321644 | 11.30 |
ENSRNOT00000075171
|
Tbc1d16
|
TBC1 domain family, member 16 |
| chr15_+_32811135 | 11.27 |
ENSRNOT00000067689
|
AABR07017902.1
|
|
| chr11_-_76888178 | 11.21 |
ENSRNOT00000049841
|
Ostn
|
osteocrin |
| chr10_-_88163712 | 11.19 |
ENSRNOT00000005382
ENSRNOT00000084493 |
Krt17
|
keratin 17 |
| chr19_+_568287 | 11.05 |
ENSRNOT00000016419
|
Cdh16
|
cadherin 16 |
| chr6_-_143195143 | 10.92 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chrX_-_38196060 | 10.87 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr17_-_15478343 | 10.86 |
ENSRNOT00000078029
|
Omd
|
osteomodulin |
| chr17_-_14637832 | 10.80 |
ENSRNOT00000074323
|
LOC103690116
|
osteomodulin |
| chrX_-_31851715 | 10.76 |
ENSRNOT00000068601
|
Vegfd
|
vascular endothelial growth factor D |
| chr2_-_235177275 | 10.72 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chrX_+_65226748 | 10.33 |
ENSRNOT00000076181
|
Msn
|
moesin |
| chr17_-_81187739 | 10.04 |
ENSRNOT00000063911
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr10_-_88122233 | 10.00 |
ENSRNOT00000083895
ENSRNOT00000005285 |
Krt14
|
keratin 14 |
| chr1_+_218466289 | 9.95 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr2_+_68820615 | 9.94 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr11_+_42945084 | 9.80 |
ENSRNOT00000002292
|
Crybg3
|
crystallin beta-gamma domain containing 3 |
| chr15_+_104026601 | 9.79 |
ENSRNOT00000013520
ENSRNOT00000083269 ENSRNOT00000093403 |
Cldn10
|
claudin 10 |
| chr8_+_99880073 | 9.73 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr12_-_5685448 | 9.70 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr3_+_148541909 | 9.62 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr2_+_68821004 | 9.55 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr3_+_19045214 | 9.40 |
ENSRNOT00000070878
|
AABR07051670.1
|
|
| chr13_+_27465930 | 9.37 |
ENSRNOT00000003314
|
Serpinb10
|
serpin family B member 10 |
| chr3_-_47025128 | 9.34 |
ENSRNOT00000011682
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_+_19495628 | 9.27 |
ENSRNOT00000077990
|
Igkv4-57
|
immunoglobulin kappa variable 4-57 |
| chr11_-_60547201 | 9.21 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr4_-_113610243 | 9.16 |
ENSRNOT00000008813
|
Hk2
|
hexokinase 2 |
| chr2_+_150756185 | 9.06 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr5_-_24631679 | 9.04 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr13_+_90116843 | 8.91 |
ENSRNOT00000006306
|
Cd48
|
Cd48 molecule |
| chr11_-_32550539 | 8.87 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr13_-_67688477 | 8.65 |
ENSRNOT00000068148
|
Prg4
|
proteoglycan 4 |
| chr11_+_71872830 | 8.62 |
ENSRNOT00000050364
|
Nrros
|
negative regulator of reactive oxygen species |
| chr9_+_16702460 | 8.42 |
ENSRNOT00000061432
|
Ptk7
|
protein tyrosine kinase 7 |
| chr4_-_103569159 | 8.41 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr4_-_103369224 | 8.35 |
ENSRNOT00000075709
|
AABR07061057.1
|
|
| chr6_-_143195445 | 8.30 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr14_-_72025137 | 8.26 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr10_-_47997796 | 8.21 |
ENSRNOT00000078422
|
Slc5a10
|
solute carrier family 5 member 10 |
| chr1_+_266255797 | 8.19 |
ENSRNOT00000027047
|
Trim8
|
tripartite motif-containing 8 |
| chr13_+_50434702 | 8.10 |
ENSRNOT00000032244
|
Sox13
|
SRY box 13 |
| chr20_-_27657983 | 8.03 |
ENSRNOT00000057313
|
Fam26e
|
family with sequence similarity 26, member E |
| chr3_-_46942966 | 8.02 |
ENSRNOT00000087439
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_-_153246433 | 7.93 |
ENSRNOT00000067748
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr4_-_103145058 | 7.86 |
ENSRNOT00000073076
|
AABR07061048.1
|
|
| chr13_-_67819835 | 7.79 |
ENSRNOT00000093666
|
Hmcn1
|
hemicentin 1 |
| chr17_+_44763598 | 7.75 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr11_-_60546997 | 7.75 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr7_+_95074236 | 7.67 |
ENSRNOT00000067649
|
Col14a1
|
collagen type XIV alpha 1 chain |
| chr6_-_109935533 | 7.62 |
ENSRNOT00000013516
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr9_-_115382445 | 7.56 |
ENSRNOT00000091316
|
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr18_-_6782757 | 7.55 |
ENSRNOT00000068150
|
Aqp4
|
aquaporin 4 |
| chr13_-_83202864 | 7.53 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr2_-_183031214 | 7.42 |
ENSRNOT00000013260
|
LOC679811
|
similar to RIKEN cDNA D930015E06 |
| chr1_+_63842277 | 7.42 |
ENSRNOT00000087957
ENSRNOT00000080820 |
Lilrb3a
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3A |
| chr15_+_32828165 | 7.35 |
ENSRNOT00000060253
|
AABR07017902.1
|
|
| chr2_-_193330699 | 7.12 |
ENSRNOT00000012684
|
Crct1
|
cysteine-rich C-terminal 1 |
| chr3_+_116899878 | 7.09 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr8_-_132919110 | 7.02 |
ENSRNOT00000008747
|
Xcr1
|
X-C motif chemokine receptor 1 |
| chr7_-_96464049 | 7.01 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr9_-_88534710 | 6.92 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr13_+_50434886 | 6.90 |
ENSRNOT00000076857
|
Sox13
|
SRY box 13 |
| chr18_-_6782996 | 6.77 |
ENSRNOT00000090320
|
Aqp4
|
aquaporin 4 |
| chr3_+_28627084 | 6.76 |
ENSRNOT00000049884
|
Arhgap15
|
Rho GTPase activating protein 15 |
| chr8_+_2604962 | 6.72 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr17_+_5281727 | 6.58 |
ENSRNOT00000024781
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chr1_-_24190896 | 6.57 |
ENSRNOT00000016121
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr8_-_18508971 | 6.48 |
ENSRNOT00000061339
|
Muc16
|
mucin 16, cell surface associated |
| chr1_-_77808462 | 6.47 |
ENSRNOT00000078917
ENSRNOT00000080422 |
LOC103689961
|
selenoprotein W-like |
| chr18_+_30562178 | 6.38 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr1_+_213070528 | 6.29 |
ENSRNOT00000051727
|
Olr302
|
olfactory receptor 302 |
| chr12_-_38916237 | 6.26 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr18_+_30826260 | 6.22 |
ENSRNOT00000065235
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chrX_-_29648359 | 6.14 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr10_+_83655460 | 6.12 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr3_-_171342646 | 6.01 |
ENSRNOT00000071853
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr1_-_77535681 | 5.91 |
ENSRNOT00000018559
|
Selenow
|
selenoprotein W |
| chr4_-_17594598 | 5.90 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr18_+_65155685 | 5.87 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr6_-_142178771 | 5.86 |
ENSRNOT00000071577
|
Ighv12-3
|
immunoglobulin heavy variable V12-3 |
| chr1_+_164502389 | 5.74 |
ENSRNOT00000043554
|
Arrb1
|
arrestin, beta 1 |
| chr4_+_99168248 | 5.69 |
ENSRNOT00000058186
|
LOC102552663
|
uncharacterized LOC102552663 |
| chr6_+_8284878 | 5.69 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr16_+_74810938 | 5.62 |
ENSRNOT00000058091
|
Nek5
|
NIMA-related kinase 5 |
| chr1_-_8751198 | 5.61 |
ENSRNOT00000030511
|
Adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr18_+_30496318 | 5.51 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr4_-_157433467 | 5.50 |
ENSRNOT00000028965
|
Lag3
|
lymphocyte activating 3 |
| chr5_-_58183017 | 5.47 |
ENSRNOT00000020982
|
Ccl19
|
C-C motif chemokine ligand 19 |
| chr8_-_23146689 | 5.47 |
ENSRNOT00000092200
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr3_-_48536235 | 5.46 |
ENSRNOT00000083536
|
Fap
|
fibroblast activation protein, alpha |
| chr8_-_39551700 | 5.46 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr5_+_133864798 | 5.44 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr8_+_116324040 | 5.42 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr7_-_142062870 | 5.40 |
ENSRNOT00000026531
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr10_-_46206135 | 5.36 |
ENSRNOT00000091471
|
Cops3
|
COP9 signalosome subunit 3 |
| chr2_-_208215187 | 5.31 |
ENSRNOT00000047836
|
Rap1a
|
RAP1A, member of RAS oncogene family |
| chr13_+_92241306 | 5.23 |
ENSRNOT00000004677
|
Olr1595
|
olfactory receptor 1595 |
| chr4_+_102262007 | 5.22 |
ENSRNOT00000079328
|
AABR07060992.2
|
|
| chr14_-_60276794 | 5.21 |
ENSRNOT00000048509
|
Slc34a2
|
solute carrier family 34 member 2 |
| chr14_-_84170835 | 5.13 |
ENSRNOT00000005646
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr4_+_181055662 | 5.10 |
ENSRNOT00000086813
|
Stk38l
|
serine/threonine kinase 38 like |
| chr3_-_64024205 | 5.09 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr3_+_19128400 | 5.01 |
ENSRNOT00000074272
|
AABR07051673.1
|
|
| chr1_+_88885937 | 4.97 |
ENSRNOT00000029719
|
Nfkbid
|
NFKB inhibitor delta |
| chr14_-_44225713 | 4.94 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr4_-_29778039 | 4.93 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr13_-_68360664 | 4.92 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr8_-_48850671 | 4.92 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr16_-_3912148 | 4.91 |
ENSRNOT00000014941
|
Anxa11
|
annexin A11 |
| chr14_-_81053905 | 4.89 |
ENSRNOT00000045068
ENSRNOT00000040215 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr3_+_140024043 | 4.87 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chr4_+_129574264 | 4.67 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chr16_+_75572070 | 4.61 |
ENSRNOT00000043486
|
Defb52
|
defensin beta 52 |
| chrX_+_18489597 | 4.59 |
ENSRNOT00000059744
|
LOC681355
|
similar to potassium channel tetramerisation domain containing 12b |
| chr1_-_116153722 | 4.58 |
ENSRNOT00000041605
|
Fpr3
|
formyl peptide receptor 3 |
| chr15_+_4285052 | 4.57 |
ENSRNOT00000035345
|
Cfap70
|
cilia and flagella associated protein 70 |
| chr18_+_30864216 | 4.54 |
ENSRNOT00000027015
|
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chrX_-_25590048 | 4.51 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr2_+_216863428 | 4.47 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr18_+_86299463 | 4.44 |
ENSRNOT00000058152
|
Cd226
|
CD226 molecule |
| chr7_-_142063212 | 4.40 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr1_-_228753422 | 4.29 |
ENSRNOT00000028626
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr5_-_99033107 | 4.29 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr16_-_74020437 | 4.23 |
ENSRNOT00000037389
|
Kat6a
|
lysine acetyltransferase 6A |
| chr18_+_30890869 | 4.22 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr10_+_34489940 | 4.18 |
ENSRNOT00000085975
|
Zfp62
|
zinc finger protein 62 |
| chr9_+_79659251 | 4.17 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr2_+_165601007 | 4.08 |
ENSRNOT00000013931
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr5_-_147761983 | 4.05 |
ENSRNOT00000012936
|
Lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr14_-_84170301 | 4.05 |
ENSRNOT00000080413
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr2_+_186555632 | 4.00 |
ENSRNOT00000052347
|
Fcrl1
|
Fc receptor-like 1 |
| chr6_+_115170865 | 3.98 |
ENSRNOT00000005671
|
Tshr
|
thyroid stimulating hormone receptor |
| chr18_-_74542077 | 3.98 |
ENSRNOT00000078519
|
Slc14a2
|
solute carrier family 14 member 2 |
| chrX_-_104984341 | 3.93 |
ENSRNOT00000077996
|
Xkrx
|
XK related, X-linked |
| chr13_-_77896697 | 3.88 |
ENSRNOT00000003452
|
Tnn
|
tenascin N |
| chr12_+_23544287 | 3.74 |
ENSRNOT00000001938
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr16_-_81028656 | 3.71 |
ENSRNOT00000091296
|
Dcun1d2
|
defective in cullin neddylation 1 domain containing 2 |
| chr9_-_61810417 | 3.71 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr3_+_151126591 | 3.70 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr7_+_141237768 | 3.69 |
ENSRNOT00000091717
|
Aqp2
|
aquaporin 2 |
| chr7_-_97071968 | 3.67 |
ENSRNOT00000006596
|
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr14_+_73674753 | 3.63 |
ENSRNOT00000040676
|
RGD1561694
|
similar to High mobility group protein 2 (HMG-2) |
| chr18_+_28409955 | 3.62 |
ENSRNOT00000026989
|
Paip2
|
poly(A) binding protein interacting protein 2 |
| chr4_+_56337695 | 3.62 |
ENSRNOT00000071926
|
Lep
|
leptin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.9 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 9.3 | 28.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 8.9 | 35.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 6.1 | 24.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 5.8 | 11.5 | GO:1904732 | regulation of electron carrier activity(GO:1904732) |
| 5.6 | 16.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 5.3 | 21.1 | GO:2001013 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 4.6 | 22.8 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 4.2 | 12.7 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 4.2 | 20.9 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 4.1 | 16.6 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 3.9 | 38.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 3.7 | 11.2 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 3.7 | 18.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 3.6 | 3.6 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 3.6 | 18.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 3.6 | 14.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 3.6 | 10.8 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 3.6 | 14.3 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 3.6 | 21.4 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 3.5 | 14.1 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 3.3 | 10.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 3.3 | 9.8 | GO:0035444 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 3.2 | 16.2 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 2.9 | 11.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 2.8 | 19.5 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 2.7 | 16.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 2.6 | 12.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 2.5 | 7.6 | GO:0003032 | detection of oxygen(GO:0003032) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 2.5 | 15.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 2.5 | 7.5 | GO:0071661 | immunoglobulin production in mucosal tissue(GO:0002426) T-helper 1 cell activation(GO:0035711) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) regulation of thymocyte migration(GO:2000410) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 2.5 | 7.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 2.4 | 9.7 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 2.3 | 9.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 2.1 | 19.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 2.1 | 12.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 2.1 | 10.3 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 2.0 | 6.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 2.0 | 7.9 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 2.0 | 21.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.8 | 3.7 | GO:0015793 | glycerol transport(GO:0015793) |
| 1.8 | 5.5 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 1.8 | 5.4 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 1.8 | 10.8 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.7 | 17.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 1.7 | 6.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.7 | 10.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.7 | 5.0 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 1.6 | 4.9 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 1.6 | 6.4 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 1.6 | 3.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 1.6 | 17.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 1.5 | 46.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 1.5 | 23.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 1.5 | 21.4 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 1.5 | 4.4 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.4 | 13.6 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 1.3 | 4.0 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 1.2 | 6.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 1.2 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 1.2 | 2.3 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 1.1 | 16.0 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 1.1 | 2.3 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 1.1 | 11.2 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 1.1 | 8.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 1.1 | 3.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 1.1 | 5.5 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.1 | 3.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 1.1 | 5.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 1.0 | 4.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 1.0 | 3.0 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of osteoclast proliferation(GO:0090289) |
| 1.0 | 6.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 1.0 | 3.9 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 1.0 | 29.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.9 | 6.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.9 | 8.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.9 | 3.7 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.9 | 5.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.9 | 2.7 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.8 | 26.8 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.8 | 4.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.8 | 2.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.8 | 4.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.8 | 2.3 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.8 | 6.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.7 | 5.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.7 | 8.9 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.7 | 9.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.7 | 8.9 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.7 | 17.8 | GO:0048305 | inactivation of MAPK activity(GO:0000188) immunoglobulin secretion(GO:0048305) |
| 0.6 | 4.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.6 | 1.9 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.6 | 5.6 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.6 | 3.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.6 | 11.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.6 | 5.9 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.6 | 2.9 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.6 | 1.1 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.6 | 2.8 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.5 | 7.7 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.5 | 2.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.5 | 1.6 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.5 | 11.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.5 | 4.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.5 | 5.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.5 | 3.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 1.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.4 | 3.5 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.4 | 8.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.4 | 3.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.4 | 4.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 2.4 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.4 | 1.6 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.4 | 1.5 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.4 | 1.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 9.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.3 | 2.9 | GO:0060613 | fat pad development(GO:0060613) |
| 0.3 | 13.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 7.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.3 | 0.9 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.3 | 2.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.3 | 4.9 | GO:0048535 | lymph node development(GO:0048535) |
| 0.3 | 2.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.3 | 7.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.3 | 5.9 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.3 | 1.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 8.2 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 1.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 4.2 | GO:0035909 | aorta morphogenesis(GO:0035909) |
| 0.2 | 0.9 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 2.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 5.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 13.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.2 | 3.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 0.8 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.2 | 6.7 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.2 | 5.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 0.7 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.2 | 8.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.2 | 0.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 3.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 5.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.2 | 2.6 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 1.3 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.2 | 0.8 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 13.2 | GO:0048593 | camera-type eye morphogenesis(GO:0048593) |
| 0.1 | 3.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 2.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 4.2 | GO:1900271 | regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 1.1 | GO:0014894 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 25.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 7.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 2.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 24.5 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 2.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 2.9 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 3.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.1 | 8.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 3.1 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 3.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 2.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.3 | GO:0061295 | pronephric nephron tubule development(GO:0039020) regulation of endodermal cell fate specification(GO:0042663) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) regulation of branch elongation involved in ureteric bud branching(GO:0072095) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 1.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 2.8 | GO:0051057 | positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 1.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 11.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.8 | GO:0051895 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) negative regulation of focal adhesion assembly(GO:0051895) positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of adherens junction organization(GO:1903392) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.4 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.0 | GO:0007320 | insemination(GO:0007320) |
| 0.1 | 1.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 1.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.7 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 1.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.7 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 1.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 2.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:0045657 | apoptotic chromosome condensation(GO:0030263) positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 5.4 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 2.3 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 3.5 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.9 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.6 | GO:0001824 | blastocyst development(GO:0001824) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 38.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 4.8 | 28.9 | GO:0030478 | actin cap(GO:0030478) |
| 4.2 | 12.7 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 4.0 | 16.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 3.7 | 3.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.3 | 9.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 2.0 | 6.1 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 1.9 | 17.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 1.8 | 12.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 1.5 | 4.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 1.3 | 6.7 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 1.2 | 16.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.2 | 3.5 | GO:0000811 | GINS complex(GO:0000811) |
| 1.1 | 20.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 1.0 | 4.2 | GO:0043564 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) Ku70:Ku80 complex(GO:0043564) |
| 0.8 | 30.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.8 | 5.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.7 | 2.7 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.6 | 16.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.6 | 21.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.6 | 2.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.5 | 2.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.5 | 20.0 | GO:0043034 | costamere(GO:0043034) |
| 0.5 | 17.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.5 | 111.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.5 | 4.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.5 | 3.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 8.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.5 | 2.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.5 | 9.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 3.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 4.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.4 | 5.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 4.9 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 5.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 8.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 102.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.3 | 21.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 0.8 | GO:1990032 | climbing fiber(GO:0044301) parallel fiber(GO:1990032) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 5.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 11.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 1.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 6.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 21.6 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 3.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 9.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 4.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 2.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 5.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 47.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.7 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 7.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 5.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 35.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 16.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 5.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 5.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 7.8 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 1.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 5.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.0 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 37.6 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 2.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 8.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 11.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 7.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 4.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 28.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 7.2 | 28.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 6.1 | 24.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 5.9 | 35.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 5.4 | 16.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 5.3 | 16.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 4.1 | 16.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 3.5 | 17.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 3.3 | 10.0 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 3.3 | 9.8 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) chromium ion transmembrane transporter activity(GO:0070835) |
| 3.1 | 18.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 3.0 | 9.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 2.8 | 16.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.3 | 20.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 2.2 | 8.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.1 | 8.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 2.0 | 10.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 2.0 | 5.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.9 | 5.7 | GO:0031893 | follicle-stimulating hormone receptor binding(GO:0031762) vasopressin receptor binding(GO:0031893) |
| 1.8 | 5.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.8 | 5.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.7 | 15.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.6 | 14.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 1.6 | 74.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 1.5 | 9.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.5 | 7.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.4 | 21.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 1.4 | 7.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.4 | 19.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.3 | 30.5 | GO:0015288 | porin activity(GO:0015288) |
| 1.3 | 29.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 1.3 | 8.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.3 | 26.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.1 | 3.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 1.1 | 18.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.1 | 13.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 1.0 | 5.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 1.0 | 13.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.0 | 7.9 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.9 | 8.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.8 | 3.4 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.8 | 7.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.8 | 3.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 8.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.8 | 6.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.8 | 4.6 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.8 | 6.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.8 | 2.3 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.6 | 16.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.6 | 4.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.6 | 5.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.6 | 11.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.6 | 3.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.6 | 4.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.5 | 4.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.5 | 6.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.5 | 12.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.5 | 11.8 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.4 | 17.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.4 | 48.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.4 | 2.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.4 | 8.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 7.0 | GO:0004950 | chemokine receptor activity(GO:0004950) |
| 0.4 | 7.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 2.8 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 1.3 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.3 | 1.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 2.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 3.7 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.3 | 18.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.3 | 4.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 6.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 5.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 1.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 6.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 3.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 3.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 6.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 1.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 1.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.2 | 3.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 3.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 3.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 11.5 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 3.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 2.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 1.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 2.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 11.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 5.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 3.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 2.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 5.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 9.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.8 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 4.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 11.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 4.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 6.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.2 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 5.6 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 22.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 8.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 4.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 14.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 3.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 20.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.1 | 46.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 1.0 | 52.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 1.0 | 32.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 1.0 | 28.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.0 | 19.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.9 | 29.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.9 | 32.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.8 | 26.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.7 | 26.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.7 | 6.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.6 | 24.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.6 | 19.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.5 | 28.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.4 | 31.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.4 | 12.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.4 | 5.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.4 | 53.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 14.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.3 | 29.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 1.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 5.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 1.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.3 | 15.5 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.3 | 7.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 12.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 2.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 4.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 6.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 2.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 5.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 26.4 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.1 | 2.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 3.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 4.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 4.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 2.7 | 38.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.5 | 18.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.4 | 52.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 1.1 | 31.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 1.0 | 28.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.9 | 16.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.8 | 14.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.8 | 27.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.7 | 13.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.6 | 10.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.6 | 11.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.6 | 6.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.5 | 25.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.5 | 4.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.5 | 12.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 6.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 4.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 8.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.4 | 24.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.4 | 4.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 10.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 3.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 5.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 7.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 17.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 3.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 2.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.3 | 15.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 3.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 6.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 2.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 16.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 9.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 2.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 4.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 6.1 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.1 | 1.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 24.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 7.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 4.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 7.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 5.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 11.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 6.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 4.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |