Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
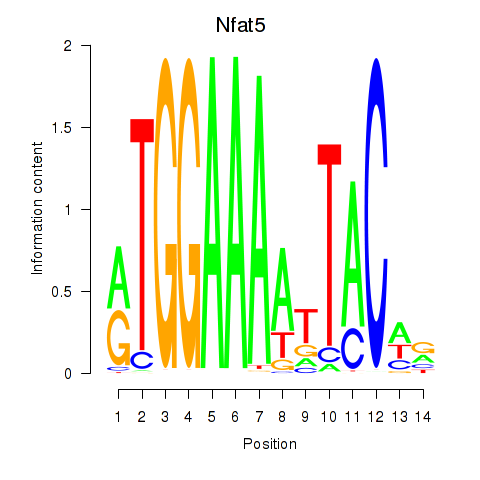
Results for Nfat5
Z-value: 1.22
Transcription factors associated with Nfat5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfat5
|
ENSRNOG00000011879 | nuclear factor of activated T-cells 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfat5 | rn6_v1_chr19_-_38533735_38533735 | 0.07 | 1.9e-01 | Click! |
Activity profile of Nfat5 motif
Sorted Z-values of Nfat5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_141237768 | 54.29 |
ENSRNOT00000091717
|
Aqp2
|
aquaporin 2 |
| chr2_+_68821004 | 50.46 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr2_+_68820615 | 48.75 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr2_-_235177275 | 47.62 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr8_+_49713190 | 42.36 |
ENSRNOT00000022074
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr9_+_95501778 | 42.28 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr18_-_69944632 | 39.40 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr10_+_71159869 | 35.72 |
ENSRNOT00000075977
ENSRNOT00000047427 |
Hnf1b
|
HNF1 homeobox B |
| chr9_+_95161157 | 30.91 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr4_-_177196180 | 28.99 |
ENSRNOT00000018999
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr3_-_94808861 | 28.89 |
ENSRNOT00000038464
|
Prrg4
|
proline rich and Gla domain 4 |
| chr6_+_8284878 | 24.85 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr5_-_146795866 | 24.63 |
ENSRNOT00000065640
|
Tlr12
|
toll-like receptor 12 |
| chr10_+_109665682 | 22.34 |
ENSRNOT00000054963
|
Slc25a10
|
solute carrier family 25 member 10 |
| chr12_-_29743705 | 21.93 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr7_-_68549763 | 21.82 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr7_-_97071968 | 21.33 |
ENSRNOT00000006596
|
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr9_-_60255746 | 17.15 |
ENSRNOT00000093710
|
Dnah7
|
dynein, axonemal, heavy chain 7 |
| chr4_+_98337367 | 16.94 |
ENSRNOT00000042165
|
AABR07060872.1
|
|
| chr5_-_160620334 | 16.93 |
ENSRNOT00000019171
|
Tmem51
|
transmembrane protein 51 |
| chr15_+_39779648 | 16.80 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr1_+_256382791 | 15.89 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr1_+_213886775 | 15.75 |
ENSRNOT00000020864
|
Pkp3
|
plakophilin 3 |
| chr6_-_42616548 | 15.26 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr3_-_148057523 | 14.68 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr7_-_27552078 | 13.72 |
ENSRNOT00000059538
|
Stab2
|
stabilin 2 |
| chr6_-_107080524 | 13.51 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr3_-_153321352 | 13.38 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr9_-_60330086 | 12.83 |
ENSRNOT00000092162
ENSRNOT00000093630 |
Dnah7
|
dynein, axonemal, heavy chain 7 |
| chr9_+_30939555 | 12.66 |
ENSRNOT00000016887
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr4_-_123713319 | 12.56 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr16_+_54765325 | 12.41 |
ENSRNOT00000065327
ENSRNOT00000086899 |
Mtmr7
|
myotubularin related protein 7 |
| chrX_+_43626480 | 12.31 |
ENSRNOT00000068078
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr5_+_135351816 | 12.30 |
ENSRNOT00000082602
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chr1_-_24190896 | 12.22 |
ENSRNOT00000016121
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_143195445 | 11.87 |
ENSRNOT00000078672
|
AABR07065837.1
|
|
| chr14_+_48726045 | 11.70 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr8_+_5967463 | 11.01 |
ENSRNOT00000086251
|
Tmem123
|
transmembrane protein 123 |
| chr8_-_128672284 | 11.00 |
ENSRNOT00000083004
|
Csrnp1
|
cysteine and serine rich nuclear protein 1 |
| chr11_-_11585765 | 10.80 |
ENSRNOT00000066439
|
Robo2
|
roundabout guidance receptor 2 |
| chr1_+_41192824 | 10.53 |
ENSRNOT00000082133
|
Esr1
|
estrogen receptor 1 |
| chr2_-_28801223 | 10.46 |
ENSRNOT00000020726
|
Tmem171
|
transmembrane protein 171 |
| chr11_+_32211115 | 10.41 |
ENSRNOT00000087452
|
Mrps6
|
mitochondrial ribosomal protein S6 |
| chr2_-_112802073 | 10.26 |
ENSRNOT00000093081
|
Ect2
|
epithelial cell transforming 2 |
| chr6_-_143195143 | 9.98 |
ENSRNOT00000081337
|
AABR07065837.1
|
|
| chr2_+_185590986 | 9.78 |
ENSRNOT00000088807
ENSRNOT00000088188 |
Lrba
|
LPS responsive beige-like anchor protein |
| chr16_+_49350884 | 9.44 |
ENSRNOT00000014828
|
Snx25
|
sorting nexin 25 |
| chrX_+_110894845 | 9.41 |
ENSRNOT00000080072
|
Tbc1d8b
|
TBC1 domain family member 8B |
| chr5_-_100970043 | 9.20 |
ENSRNOT00000013950
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr16_+_74534130 | 8.88 |
ENSRNOT00000050637
|
Slc25a15
|
solute carrier family 25 member 15 |
| chr1_-_194769524 | 8.78 |
ENSRNOT00000025988
|
Nupr1
|
nuclear protein 1, transcriptional regulator |
| chrX_-_82699487 | 8.60 |
ENSRNOT00000081625
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr1_-_198045154 | 7.99 |
ENSRNOT00000072875
|
Nupr1l1
|
nuclear protein, transcriptional regulator, 1-like 1 |
| chr2_+_165601007 | 7.66 |
ENSRNOT00000013931
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr18_+_25749098 | 7.66 |
ENSRNOT00000027771
|
Camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr4_-_178441547 | 7.66 |
ENSRNOT00000087434
ENSRNOT00000055542 |
Sox5
|
SRY box 5 |
| chr3_-_2490392 | 7.44 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr10_+_108132105 | 7.34 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr16_-_82288022 | 7.34 |
ENSRNOT00000078609
|
Spaca7
|
sperm acrosome associated 7 |
| chr13_-_78521911 | 6.82 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr7_+_74350479 | 6.75 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr17_+_24242642 | 6.65 |
ENSRNOT00000024323
|
Rnf182
|
ring finger protein 182 |
| chr6_+_126766967 | 6.42 |
ENSRNOT00000052296
|
Cox8c
|
cytochrome c oxidase subunit 8C |
| chr12_+_37702404 | 6.29 |
ENSRNOT00000049985
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr15_+_10120206 | 6.08 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr2_+_180012414 | 5.90 |
ENSRNOT00000082273
|
Pdgfc
|
platelet derived growth factor C |
| chr5_-_153924896 | 5.80 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr3_-_77483322 | 5.50 |
ENSRNOT00000008077
|
Olr661
|
olfactory receptor 661 |
| chr15_+_5904569 | 5.41 |
ENSRNOT00000072599
|
LOC102546376
|
disks large homolog 5-like |
| chr7_+_20262680 | 4.98 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr16_-_81693000 | 4.92 |
ENSRNOT00000092353
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr13_+_49003803 | 4.81 |
ENSRNOT00000080556
|
Lemd1
|
LEM domain containing 1 |
| chr12_+_21767606 | 4.79 |
ENSRNOT00000059602
|
LOC103691013
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr8_-_6305033 | 4.66 |
ENSRNOT00000029887
|
Cep126
|
centrosomal protein 126 |
| chr18_+_29386809 | 4.36 |
ENSRNOT00000082079
ENSRNOT00000024825 |
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr11_+_47243342 | 4.31 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr14_+_81819799 | 4.22 |
ENSRNOT00000076840
|
Mxd4
|
Max dimerization protein 4 |
| chr1_-_211196868 | 4.19 |
ENSRNOT00000022714
|
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr10_-_73629581 | 4.11 |
ENSRNOT00000091172
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr16_-_75107931 | 3.88 |
ENSRNOT00000058066
|
Defb15
|
defensin beta 15 |
| chr1_+_199655660 | 3.78 |
ENSRNOT00000027032
|
Armc5
|
armadillo repeat containing 5 |
| chr7_-_140556639 | 3.70 |
ENSRNOT00000089523
|
Rhebl1
|
Ras homolog enriched in brain like 1 |
| chr12_+_23544287 | 3.59 |
ENSRNOT00000001938
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr20_-_3372070 | 3.40 |
ENSRNOT00000080770
|
Dhx16
|
DEAH-box helicase 16 |
| chr5_+_64566804 | 3.34 |
ENSRNOT00000073192
|
LOC103690035
|
TGF-beta receptor type-1-like |
| chr16_+_75122996 | 3.23 |
ENSRNOT00000018375
|
Defb15
|
defensin beta 15 |
| chr16_-_75518336 | 2.95 |
ENSRNOT00000058037
|
Defa11
|
defensin alpha 11 |
| chr5_-_24926904 | 2.95 |
ENSRNOT00000075288
|
AABR07047232.1
|
|
| chr12_-_41590244 | 2.86 |
ENSRNOT00000038736
|
Slc8b1
|
solute carrier family 8 member B1 |
| chr12_-_18227991 | 2.82 |
ENSRNOT00000072986
|
AABR07035501.1
|
|
| chr2_+_87418517 | 2.74 |
ENSRNOT00000048046
|
LOC100909879
|
tyrosine-protein phosphatase non-receptor type substrate 1-like |
| chr19_-_52424318 | 2.49 |
ENSRNOT00000021743
|
Tldc1
|
TBC/LysM-associated domain containing 1 |
| chr5_+_130286393 | 2.48 |
ENSRNOT00000044795
|
Ccp6l1
|
cytosolic carboxypeptidase 6-like 1 |
| chr1_-_274107138 | 2.43 |
ENSRNOT00000078670
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr13_-_27653839 | 2.23 |
ENSRNOT00000071638
|
LOC100360575
|
signal-regulatory protein alpha-like |
| chr7_+_23012070 | 2.03 |
ENSRNOT00000042460
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr9_+_37462693 | 1.99 |
ENSRNOT00000016316
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr10_-_87407634 | 1.93 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr6_-_98566523 | 1.90 |
ENSRNOT00000077715
|
Ppp2r5e
|
protein phosphatase 2 regulatory subunit B', epsilon |
| chr10_+_83081168 | 1.89 |
ENSRNOT00000035023
|
Tac4
|
tachykinin 4 (hemokinin) |
| chr7_-_15821927 | 1.85 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr14_-_78902063 | 1.77 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr20_+_1936438 | 1.69 |
ENSRNOT00000073981
|
Olr1749
|
olfactory receptor 1749 |
| chr1_+_57345577 | 1.53 |
ENSRNOT00000082790
ENSRNOT00000084017 |
Fam120b
|
family with sequence similarity 120B |
| chr15_-_61648267 | 1.45 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr11_+_43340505 | 1.02 |
ENSRNOT00000043299
|
Olr1541
|
olfactory receptor 1541 |
| chr8_+_132120452 | 1.00 |
ENSRNOT00000055822
|
Tgm4
|
transglutaminase 4 |
| chr6_+_73358112 | 0.79 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr15_+_70790125 | 0.75 |
ENSRNOT00000012440
|
Tdrd3
|
tudor domain containing 3 |
| chr8_+_22423467 | 0.69 |
ENSRNOT00000085320
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr19_-_59894937 | 0.39 |
ENSRNOT00000027145
|
Rbm34
|
RNA binding motif protein 34 |
| chr18_+_60745035 | 0.37 |
ENSRNOT00000045098
|
LOC502176
|
hypothetical protein LOC502176 |
| chr3_+_2490518 | 0.35 |
ENSRNOT00000015099
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr7_-_134495835 | 0.26 |
ENSRNOT00000058349
|
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr7_-_15723127 | 0.13 |
ENSRNOT00000071599
|
LOC100909683
|
olfactory receptor 63-like |
| chr8_-_76940094 | 0.11 |
ENSRNOT00000082709
ENSRNOT00000084313 |
Rnf111
|
ring finger protein 111 |
| chr10_-_25847994 | 0.11 |
ENSRNOT00000082076
|
Mat2b
|
methionine adenosyltransferase 2B |
| chrX_-_118459355 | 0.03 |
ENSRNOT00000084465
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr8_-_42944070 | 0.01 |
ENSRNOT00000075290
|
LOC100910409
|
olfactory receptor 146-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfat5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.1 | 54.3 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 12.4 | 99.2 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 11.9 | 35.7 | GO:0061228 | mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 7.4 | 22.3 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 5.3 | 21.3 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 4.4 | 21.8 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 4.0 | 15.9 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 3.4 | 30.9 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 3.1 | 15.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 3.1 | 12.3 | GO:0032919 | spermine acetylation(GO:0032919) |
| 2.6 | 10.5 | GO:1990375 | baculum development(GO:1990375) |
| 2.5 | 42.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 2.2 | 10.8 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.9 | 7.4 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 1.8 | 8.9 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 1.8 | 30.0 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 1.6 | 4.9 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.5 | 7.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.5 | 7.7 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.4 | 12.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 1.3 | 29.0 | GO:0097503 | sialylation(GO:0097503) |
| 1.1 | 9.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.9 | 24.6 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.8 | 12.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.8 | 4.1 | GO:1990918 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.8 | 6.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.7 | 12.7 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.7 | 9.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.7 | 8.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.6 | 10.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.6 | 7.7 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.6 | 8.8 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.5 | 6.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 5.9 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.4 | 11.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 42.3 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.4 | 5.8 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 1.9 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.3 | 12.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.3 | 8.2 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 13.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.3 | 12.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 16.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 4.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 3.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 1.0 | GO:0042628 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.2 | 11.0 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.2 | 13.7 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.2 | 3.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 13.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 21.9 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 1.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 2.5 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.1 | 2.0 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.1 | 1.5 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.1 | 4.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 39.4 | GO:0000165 | MAPK cascade(GO:0000165) |
| 0.1 | 7.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 3.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.8 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 13.0 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 4.4 | GO:0045137 | development of primary sexual characteristics(GO:0045137) |
| 0.0 | 2.9 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.0 | GO:0043305 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of leukocyte degranulation(GO:0043301) negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 4.3 | GO:0006954 | inflammatory response(GO:0006954) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 30.0 | GO:0036156 | inner dynein arm(GO:0036156) |
| 4.5 | 13.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 3.5 | 42.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 3.1 | 15.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 2.6 | 10.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.5 | 54.3 | GO:0046930 | pore complex(GO:0046930) |
| 1.2 | 4.9 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 1.1 | 10.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.0 | 7.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.8 | 21.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 15.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 25.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.5 | 7.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.4 | 6.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 12.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.4 | 10.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 24.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 99.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 1.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 2.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 12.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 53.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 16.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 24.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 21.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 6.3 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 7.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 7.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 9.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 8.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 4.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 6.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 21.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 9.4 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 12.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 5.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.8 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 54.3 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 7.4 | 22.3 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 5.8 | 99.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 5.3 | 15.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 4.4 | 21.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 4.2 | 12.6 | GO:0030977 | taurine binding(GO:0030977) |
| 4.1 | 12.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 3.0 | 42.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 2.1 | 10.5 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 1.8 | 21.3 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 1.7 | 12.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.7 | 15.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.6 | 12.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.4 | 39.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 1.3 | 29.0 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 1.2 | 13.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 1.2 | 15.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.1 | 8.9 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 1.0 | 30.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.9 | 10.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.8 | 9.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.6 | 7.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 13.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.5 | 6.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 12.4 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.4 | 3.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.4 | 13.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 4.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 11.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 30.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 9.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.3 | 34.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.3 | 5.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 6.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 3.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 10.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 60.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 40.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 4.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 5.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 19.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 8.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.0 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 8.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.5 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 6.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 7.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 2.7 | GO:0003712 | transcription cofactor activity(GO:0003712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 99.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.6 | 35.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 18.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.4 | 12.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 11.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 12.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 17.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 6.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 7.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 9.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 5.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 99.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 4.5 | 54.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.6 | 30.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.6 | 21.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.1 | 12.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 1.1 | 13.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.1 | 16.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 1.0 | 59.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.9 | 35.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.9 | 42.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.7 | 12.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.6 | 15.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.5 | 9.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.5 | 13.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 15.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.5 | 5.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 16.3 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.2 | 10.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 10.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 10.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 4.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 9.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 11.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 7.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |