Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
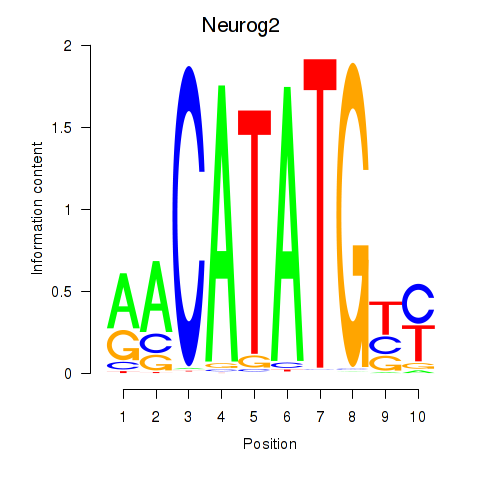
Results for Neurog2
Z-value: 0.75
Transcription factors associated with Neurog2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurog2
|
ENSRNOG00000010972 | neurogenin 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurog2 | rn6_v1_chr2_+_231962517_231962517 | -0.17 | 2.6e-03 | Click! |
Activity profile of Neurog2 motif
Sorted Z-values of Neurog2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_128003418 | 45.35 |
ENSRNOT00000013896
|
Serpina3c
|
serine (or cysteine) proteinase inhibitor, clade A, member 3C |
| chr6_+_128048099 | 38.08 |
ENSRNOT00000084685
ENSRNOT00000087017 |
LOC500712
|
Ab1-233 |
| chr1_+_148240504 | 27.80 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr15_+_28028521 | 26.13 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr19_+_42097995 | 25.00 |
ENSRNOT00000020197
|
Hp
|
haptoglobin |
| chr5_-_78985990 | 24.82 |
ENSRNOT00000009248
|
Ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr4_-_31730386 | 23.81 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr3_-_101474890 | 23.16 |
ENSRNOT00000091869
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr7_+_28414350 | 21.05 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr8_-_111777602 | 20.99 |
ENSRNOT00000052317
ENSRNOT00000083855 |
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr7_-_2431197 | 19.88 |
ENSRNOT00000003498
|
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr19_-_22632071 | 18.45 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr1_+_145715969 | 16.55 |
ENSRNOT00000037996
|
Tmc3
|
transmembrane channel-like 3 |
| chr11_-_88972176 | 16.22 |
ENSRNOT00000002498
|
Pkp2
|
plakophilin 2 |
| chr1_+_72810545 | 16.02 |
ENSRNOT00000092117
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr17_+_30556884 | 15.87 |
ENSRNOT00000080929
ENSRNOT00000022022 |
Eci2
|
enoyl-CoA delta isomerase 2 |
| chr17_+_69468427 | 15.69 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr4_+_122365093 | 14.93 |
ENSRNOT00000024011
|
Klf15
|
Kruppel-like factor 15 |
| chr1_-_205630073 | 13.80 |
ENSRNOT00000037064
|
Tex36
|
testis expressed 36 |
| chr3_+_55910177 | 12.78 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr20_-_5754773 | 12.71 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr5_-_64946580 | 12.67 |
ENSRNOT00000007290
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr9_+_95285592 | 10.76 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr3_-_148057523 | 10.54 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr4_-_82194927 | 10.48 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr1_-_213534641 | 10.19 |
ENSRNOT00000033074
|
Syce1
|
synaptonemal complex central element protein 1 |
| chr5_-_119564846 | 10.02 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr17_-_51912496 | 9.65 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr16_-_75581325 | 9.63 |
ENSRNOT00000058031
|
Defb33
|
defensin beta 33 |
| chrX_+_110342718 | 8.92 |
ENSRNOT00000043861
|
AABR07040792.1
|
|
| chr18_+_70739492 | 8.88 |
ENSRNOT00000085601
|
Acaa2
|
acetyl-CoA acyltransferase 2 |
| chr6_-_110681408 | 8.83 |
ENSRNOT00000078265
|
Angel1
|
angel homolog 1 |
| chr20_+_3677474 | 8.77 |
ENSRNOT00000047663
|
Mt1m
|
metallothionein 1M |
| chr7_+_71065197 | 8.75 |
ENSRNOT00000051075
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr2_-_100372252 | 8.72 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr4_+_79868442 | 8.61 |
ENSRNOT00000089973
|
Mpp6
|
membrane palmitoylated protein 6 |
| chr20_-_29579578 | 8.45 |
ENSRNOT00000000700
|
AC096404.1
|
|
| chrX_-_130794673 | 8.19 |
ENSRNOT00000080612
|
LOC691030
|
similar to RIKEN cDNA 1700001F22 |
| chr11_+_54137639 | 8.14 |
ENSRNOT00000066343
|
LOC100909977
|
leukocyte surface antigen CD47-like |
| chr2_-_184993341 | 7.78 |
ENSRNOT00000071580
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr1_+_129613497 | 7.73 |
ENSRNOT00000074410
|
LOC102551255
|
tetraspanin-7-like |
| chr1_-_189181901 | 7.63 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr12_-_18227991 | 7.56 |
ENSRNOT00000072986
|
AABR07035501.1
|
|
| chr8_+_72743426 | 7.34 |
ENSRNOT00000072573
|
Rps27l
|
ribosomal protein S27-like |
| chr3_+_15788778 | 7.25 |
ENSRNOT00000041240
|
Olr397
|
olfactory receptor 397 |
| chr10_+_94192556 | 7.15 |
ENSRNOT00000074007
ENSRNOT00000093039 |
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr2_-_252505971 | 7.12 |
ENSRNOT00000021984
|
LOC56764
|
dnaj-like protein |
| chr10_+_65772443 | 7.07 |
ENSRNOT00000013296
|
Sebox
|
SEBOX homeobox |
| chr14_+_2325308 | 7.07 |
ENSRNOT00000000072
|
Atp5i
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr19_+_683484 | 6.98 |
ENSRNOT00000090332
|
Terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr15_-_5486590 | 6.96 |
ENSRNOT00000040447
|
AABR07016950.1
|
|
| chr16_+_71889235 | 6.83 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr3_-_146407252 | 6.82 |
ENSRNOT00000091310
|
Apmap
|
adipocyte plasma membrane associated protein |
| chr8_-_43024557 | 6.53 |
ENSRNOT00000073494
|
Olr1273
|
olfactory receptor 1273 |
| chr10_-_40489808 | 6.49 |
ENSRNOT00000037274
|
Slc36a3
|
solute carrier family 36, member 3 |
| chr20_-_1099336 | 6.32 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chr13_-_100189778 | 6.32 |
ENSRNOT00000071062
|
LOC100912759
|
uncharacterized LOC100912759 |
| chr10_-_56530842 | 6.31 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chrX_+_122313470 | 6.27 |
ENSRNOT00000018322
|
RGD1566387
|
similar to hypothetical protein |
| chr2_+_21931493 | 6.17 |
ENSRNOT00000018259
|
Dhfr
|
dihydrofolate reductase |
| chr1_-_154507039 | 5.99 |
ENSRNOT00000078883
|
Ccdc83
|
coiled-coil domain containing 83 |
| chr4_-_22424862 | 5.87 |
ENSRNOT00000082359
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr13_-_52834134 | 5.84 |
ENSRNOT00000038874
|
Igfn1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr6_-_127571016 | 5.84 |
ENSRNOT00000057322
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chrX_-_123894411 | 5.82 |
ENSRNOT00000034083
|
Rhox2
|
reproductive homeobox on X chromosome 2 |
| chr4_-_82280722 | 5.79 |
ENSRNOT00000071668
|
Hoxa9
|
homeobox A9 |
| chr6_-_62798384 | 5.75 |
ENSRNOT00000093200
|
Ndufa10l1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10-like 1 |
| chr11_+_55474820 | 5.73 |
ENSRNOT00000040655
|
Olr1453
|
olfactory receptor 1453 |
| chr13_+_49005405 | 5.50 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr19_-_43215281 | 5.42 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr5_+_24725821 | 5.40 |
ENSRNOT00000061413
|
AABR07047221.1
|
|
| chr8_+_117920944 | 5.38 |
ENSRNOT00000028125
|
Nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr10_+_69737328 | 5.23 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr2_+_196334626 | 5.21 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr10_-_91581232 | 5.15 |
ENSRNOT00000066453
ENSRNOT00000068686 |
AABR07030520.1
|
|
| chr3_+_138174054 | 5.07 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr10_-_12491068 | 5.01 |
ENSRNOT00000071158
|
Olr1368
|
olfactory receptor 1368 |
| chr4_-_173640684 | 5.01 |
ENSRNOT00000010728
|
Rergl
|
RERG like |
| chr20_+_33527932 | 4.99 |
ENSRNOT00000066492
|
Nepn
|
nephrocan |
| chr2_+_115225356 | 4.95 |
ENSRNOT00000073329
|
LOC100910086
|
olfactory receptor 149-like |
| chr2_-_222935973 | 4.91 |
ENSRNOT00000071934
|
AABR07013095.1
|
|
| chr10_-_60006037 | 4.81 |
ENSRNOT00000065541
|
Olr1485
|
olfactory receptor 1485 |
| chr2_+_115290243 | 4.76 |
ENSRNOT00000074802
|
LOC100910153
|
olfactory receptor 149-like |
| chr10_-_12236107 | 4.75 |
ENSRNOT00000049390
|
Olr1381
|
olfactory receptor 1381 |
| chr5_-_134905588 | 4.74 |
ENSRNOT00000016013
|
Nsun4
|
NOP2/Sun RNA methyltransferase family member 4 |
| chr18_+_40883224 | 4.71 |
ENSRNOT00000039733
|
Lvrn
|
laeverin |
| chr1_-_8885967 | 4.67 |
ENSRNOT00000016102
|
Gje1
|
gap junction protein, epsilon 1 |
| chr6_-_139973811 | 4.66 |
ENSRNOT00000082875
|
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr15_+_35761388 | 4.54 |
ENSRNOT00000086118
|
Olr1286
|
olfactory receptor 1286 |
| chr8_-_84522588 | 4.52 |
ENSRNOT00000076213
|
Mlip
|
muscular LMNA-interacting protein |
| chr5_+_98998163 | 4.48 |
ENSRNOT00000075600
|
Pramef5
|
PRAME family member 5 |
| chr10_-_12370656 | 4.47 |
ENSRNOT00000060995
|
Olr1362
|
olfactory receptor 1362 |
| chr6_+_93385457 | 4.44 |
ENSRNOT00000010300
|
Actr10
|
actin-related protein 10 homolog |
| chr4_-_165787668 | 4.40 |
ENSRNOT00000044029
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr10_+_12100133 | 4.31 |
ENSRNOT00000060987
|
Olr1358
|
olfactory receptor 1358 |
| chr2_-_28801223 | 4.31 |
ENSRNOT00000020726
|
Tmem171
|
transmembrane protein 171 |
| chr9_-_105655471 | 4.26 |
ENSRNOT00000014921
|
RGD1560925
|
similar to 2610034M16Rik protein |
| chr8_+_42833019 | 4.25 |
ENSRNOT00000071050
|
LOC100910227
|
olfactory receptor 149-like |
| chr15_-_109316953 | 4.24 |
ENSRNOT00000019158
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr15_+_27400717 | 4.23 |
ENSRNOT00000047008
|
Olr1620
|
olfactory receptor 1620 |
| chr11_+_55497234 | 4.17 |
ENSRNOT00000070882
|
LOC108348268
|
olfactory receptor 2T33-like |
| chr15_+_5772679 | 4.07 |
ENSRNOT00000043553
|
AABR07016992.1
|
|
| chr10_+_37582706 | 4.01 |
ENSRNOT00000007630
|
RGD1306484
|
similar to growth and transformation-dependent protein |
| chr2_-_35192515 | 4.00 |
ENSRNOT00000086631
|
LOC100909937
|
olfactory receptor 149-like |
| chr8_+_41954140 | 3.98 |
ENSRNOT00000049709
|
Olr1223
|
olfactory receptor 1223 |
| chr11_-_28940641 | 3.93 |
ENSRNOT00000061605
|
Krtap22-2
|
keratin associated protein 22-2 |
| chr3_-_144321602 | 3.90 |
ENSRNOT00000049508
|
LOC689220
|
similar to Cystatin S precursor (LM protein) |
| chr16_-_75028977 | 3.78 |
ENSRNOT00000058071
|
Defb13
|
defensin beta 13 |
| chr1_+_91005261 | 3.76 |
ENSRNOT00000081282
|
Wdr62
|
WD repeat domain 62 |
| chr8_+_43408635 | 3.75 |
ENSRNOT00000043988
|
Olr1313
|
olfactory receptor 1313 |
| chr4_+_72272809 | 3.73 |
ENSRNOT00000048987
|
Olr808
|
olfactory receptor 808 |
| chr8_-_43304560 | 3.70 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr11_+_28724341 | 3.62 |
ENSRNOT00000049858
|
LOC100363287
|
RIKEN cDNA 2310034C09-like |
| chr1_-_238381871 | 3.60 |
ENSRNOT00000077601
|
Tmc1
|
transmembrane channel-like 1 |
| chr1_+_74770009 | 3.55 |
ENSRNOT00000012225
|
AABR07002473.1
|
|
| chr10_-_12644292 | 3.55 |
ENSRNOT00000041612
|
Olr1375
|
olfactory receptor 1375 |
| chr20_+_11612383 | 3.51 |
ENSRNOT00000045125
|
AC108323.1
|
|
| chr3_-_20518105 | 3.46 |
ENSRNOT00000050435
|
AABR07051735.1
|
|
| chr11_-_30300139 | 3.43 |
ENSRNOT00000079267
|
AABR07033612.1
|
|
| chr7_+_142756312 | 3.40 |
ENSRNOT00000071298
|
Ankrd33
|
ankyrin repeat domain 33 |
| chr10_+_12332142 | 3.40 |
ENSRNOT00000071686
|
Olr1361
|
olfactory receptor 1361 |
| chr5_+_141530211 | 3.38 |
ENSRNOT00000056546
|
4933427I04Rik
|
Riken cDNA 4933427I04 gene |
| chr3_+_73335149 | 3.35 |
ENSRNOT00000042537
|
Olr470
|
olfactory receptor 470 |
| chr17_-_10208360 | 3.34 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr14_+_7026769 | 3.33 |
ENSRNOT00000071955
|
LOC100359907
|
secretory calcium-binding phosphoprotein proline-glutamine rich 1-like |
| chr4_-_121969446 | 3.33 |
ENSRNOT00000085001
|
Vom1r99
|
vomeronasal 1 receptor 99 |
| chr7_+_13378338 | 3.30 |
ENSRNOT00000042747
|
Olr1073
|
olfactory receptor 1073 |
| chr20_+_649582 | 3.27 |
ENSRNOT00000074145
|
Olr1687
|
olfactory receptor 1687 |
| chr4_+_7257752 | 3.27 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr15_-_12889102 | 3.27 |
ENSRNOT00000061214
ENSRNOT00000084687 |
RGD1306063
|
similar to HT021 |
| chr10_-_12731897 | 3.27 |
ENSRNOT00000042582
|
Olr1378
|
olfactory receptor 1378 |
| chr1_+_230056710 | 3.27 |
ENSRNOT00000073539
|
Olr357
|
olfactory receptor 357 |
| chr10_-_12522230 | 3.25 |
ENSRNOT00000073401
|
Olr1370
|
olfactory receptor 1370 |
| chr3_+_102471249 | 3.25 |
ENSRNOT00000051083
|
Olr749
|
olfactory receptor 749 |
| chr15_+_27438853 | 3.25 |
ENSRNOT00000011636
|
Olr1622
|
olfactory receptor 1622 |
| chr2_+_25007218 | 3.23 |
ENSRNOT00000078413
|
LOC100912266
|
uncharacterized LOC100912266 |
| chr11_-_28736000 | 3.21 |
ENSRNOT00000064058
|
Krtap14l
|
keratin associated protein 14 like |
| chr10_-_60126696 | 3.17 |
ENSRNOT00000026857
|
Olr1485
|
olfactory receptor 1485 |
| chr11_+_28739219 | 3.13 |
ENSRNOT00000061620
|
Krtap13-2
|
keratin associated protein 13-2 |
| chr18_-_27206777 | 3.12 |
ENSRNOT00000090569
|
AABR07031691.1
|
|
| chr8_-_43443531 | 3.11 |
ENSRNOT00000007901
|
LOC103693060
|
olfactory receptor 148-like |
| chrX_-_138435391 | 3.09 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr15_-_46223385 | 3.07 |
ENSRNOT00000059211
|
Defb30
|
defensin beta 30 |
| chr5_-_134382794 | 3.06 |
ENSRNOT00000016714
|
LOC103690007
|
5-methylcytosine rRNA methyltransferase NSUN4 |
| chr2_-_153140894 | 3.04 |
ENSRNOT00000075166
|
AABR07010840.1
|
|
| chr8_-_17833773 | 3.04 |
ENSRNOT00000049758
|
Olr1117
|
olfactory receptor 1117 |
| chr8_-_37349694 | 3.02 |
ENSRNOT00000030776
|
LOC100360049
|
rCG64165-like |
| chr2_+_234298839 | 3.01 |
ENSRNOT00000072216
|
Dcm5
|
Dcm5 protein |
| chr1_+_168830222 | 2.99 |
ENSRNOT00000050333
|
Olr122
|
olfactory receptor 122 |
| chr2_-_199529864 | 2.99 |
ENSRNOT00000023742
|
Olr390
|
olfactory receptor 390 |
| chr13_+_71656651 | 2.97 |
ENSRNOT00000050365
|
Zfp648
|
zinc finger protein 648 |
| chr11_-_60819249 | 2.96 |
ENSRNOT00000043917
ENSRNOT00000042447 |
Cd200r1l
|
CD200 receptor 1-like |
| chr2_+_233909142 | 2.90 |
ENSRNOT00000073197
|
Dcm5
|
Dcm5 protein |
| chr17_+_87274944 | 2.89 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr4_-_130659697 | 2.89 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr2_+_46223006 | 2.86 |
ENSRNOT00000073159
|
Olr1265
|
olfactory receptor 1265 |
| chr1_+_167719947 | 2.81 |
ENSRNOT00000024971
|
Olr43
|
olfactory receptor 43 |
| chr8_-_42793484 | 2.78 |
ENSRNOT00000072871
|
AABR07069942.1
|
|
| chr16_-_75208230 | 2.78 |
ENSRNOT00000058059
|
Defb10
|
defensin beta 10 |
| chr3_-_103447716 | 2.70 |
ENSRNOT00000074196
|
Olr791
|
olfactory receptor 791 |
| chr11_+_43194348 | 2.69 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr8_+_43355060 | 2.68 |
ENSRNOT00000043206
|
Olr1309
|
olfactory receptor 1309 |
| chr18_+_16590408 | 2.67 |
ENSRNOT00000093715
|
Mocos
|
molybdenum cofactor sulfurase |
| chr5_-_173312023 | 2.67 |
ENSRNOT00000026671
|
Tas1r3
|
taste 1 receptor member 3 |
| chr8_+_18845579 | 2.63 |
ENSRNOT00000039181
|
Olr1125
|
olfactory receptor 1125 |
| chr1_-_99955717 | 2.63 |
ENSRNOT00000079624
|
Klk1c6
|
kallikrein 1-related peptidase C6 |
| chr1_+_169208988 | 2.62 |
ENSRNOT00000050606
|
Olr140
|
olfactory receptor 140 |
| chr1_+_213258684 | 2.60 |
ENSRNOT00000091678
|
LOC103690271
|
olfactory receptor 13A1-like |
| chr5_+_173248245 | 2.57 |
ENSRNOT00000025213
|
Mrpl20
|
mitochondrial ribosomal protein L20 |
| chr1_+_229461127 | 2.46 |
ENSRNOT00000035813
|
Olr332
|
olfactory receptor 332 |
| chr15_-_27032498 | 2.45 |
ENSRNOT00000016702
|
Olr1609
|
olfactory receptor 1609 |
| chr3_+_76035198 | 2.43 |
ENSRNOT00000080503
|
Olr596
|
olfactory receptor 596 |
| chr12_+_19561347 | 2.41 |
ENSRNOT00000060025
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr12_-_35318685 | 2.41 |
ENSRNOT00000074199
|
AABR07036167.1
|
|
| chr12_-_21450204 | 2.39 |
ENSRNOT00000065866
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr8_-_43082948 | 2.37 |
ENSRNOT00000072050
|
LOC100910667
|
olfactory receptor 8G5-like |
| chr10_-_110308514 | 2.36 |
ENSRNOT00000054927
|
Tex19.2
|
testis expressed gene 19.2 |
| chr11_+_43476319 | 2.35 |
ENSRNOT00000044928
|
Olr1548
|
olfactory receptor 1548 |
| chr7_-_13531526 | 2.35 |
ENSRNOT00000048054
|
Olr1081
|
olfactory receptor 1081 |
| chr17_-_15519282 | 2.33 |
ENSRNOT00000093577
|
Aspnl1
|
asporin-like 1 |
| chr7_-_13553963 | 2.30 |
ENSRNOT00000079450
|
Olr1082
|
olfactory receptor 1082 |
| chr10_-_16046033 | 2.29 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr7_+_13673934 | 2.25 |
ENSRNOT00000048564
|
Olr1085
|
olfactory receptor 1085 |
| chr3_+_72974274 | 2.25 |
ENSRNOT00000072310
|
Olr443
|
olfactory receptor 443 |
| chr3_-_158985814 | 2.19 |
ENSRNOT00000071070
|
LOC100909423
|
olfactory receptor 150-like |
| chr10_-_12674077 | 2.18 |
ENSRNOT00000041353
|
Olr1376
|
olfactory receptor 1376 |
| chr1_-_86749728 | 2.15 |
ENSRNOT00000047766
|
LOC681871
|
similar to putative pheromone receptor (Go-VN5) |
| chr7_-_145575981 | 2.06 |
ENSRNOT00000074622
|
LOC102555445
|
16.5 kDa submandibular gland glycoprotein-like |
| chr7_+_13631975 | 2.06 |
ENSRNOT00000083548
|
Olr1084
|
olfactory receptor 1084 |
| chr10_-_88000423 | 2.06 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr16_-_14300951 | 2.04 |
ENSRNOT00000017038
|
Rgr
|
retinal G protein coupled receptor |
| chr8_+_42650438 | 2.03 |
ENSRNOT00000047107
|
LOC108350227
|
olfactory receptor 147-like |
| chr1_+_230553486 | 2.02 |
ENSRNOT00000017572
|
Olr373
|
olfactory receptor 373 |
| chr3_-_45169118 | 1.95 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr1_+_64241865 | 1.92 |
ENSRNOT00000082095
|
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr1_+_247871807 | 1.90 |
ENSRNOT00000089563
|
Mlana
|
melan-A |
| chr7_-_12429897 | 1.87 |
ENSRNOT00000020670
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr1_-_167770210 | 1.85 |
ENSRNOT00000024948
|
Olr47
|
olfactory receptor 47 |
| chr18_+_16590197 | 1.83 |
ENSRNOT00000066583
|
Mocos
|
molybdenum cofactor sulfurase |
| chr1_+_205911552 | 1.78 |
ENSRNOT00000024751
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurog2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 25.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 6.0 | 23.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 5.3 | 21.0 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 3.9 | 23.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 3.2 | 16.2 | GO:0034334 | desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) |
| 3.1 | 18.4 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 2.5 | 24.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 2.4 | 9.6 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 1.9 | 7.6 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.8 | 8.9 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 1.5 | 5.9 | GO:0033231 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 1.4 | 37.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 1.4 | 5.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 1.2 | 6.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 1.2 | 10.8 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 1.1 | 8.8 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.1 | 1.1 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 1.0 | 12.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 1.0 | 45.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.8 | 6.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.7 | 3.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.7 | 4.7 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.6 | 7.0 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.6 | 15.7 | GO:2000757 | negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.6 | 1.7 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.6 | 3.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 2.7 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 0.5 | 19.9 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.5 | 5.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.5 | 12.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.4 | 5.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 4.5 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.4 | 1.5 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.4 | 3.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.4 | 10.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.4 | 1.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.3 | 8.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 1.7 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.3 | 0.8 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 7.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 16.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 3.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 1.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 | 1.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 | 1.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.2 | 10.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 4.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 4.5 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 4.2 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 0.5 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.9 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 188.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 3.4 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.1 | 16.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 3.8 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 3.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 2.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 5.0 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 4.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.6 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 4.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.7 | GO:0051198 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 1.6 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 3.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.7 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 6.7 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 2.7 | GO:0007411 | axon guidance(GO:0007411) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 25.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 4.2 | 21.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 3.2 | 9.6 | GO:0043511 | inhibin complex(GO:0043511) |
| 1.2 | 5.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 1.0 | 12.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.8 | 15.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.8 | 10.2 | GO:0000801 | central element(GO:0000801) |
| 0.8 | 16.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.7 | 7.0 | GO:0070187 | telosome(GO:0070187) |
| 0.5 | 7.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.5 | 2.7 | GO:1903767 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 0.5 | 5.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 5.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 3.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 1.9 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 4.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 3.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 10.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 21.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 8.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 1.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 8.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 7.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 3.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 7.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 26.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 7.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 67.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 4.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.4 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 51.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 85.0 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.4 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 5.0 | 24.8 | GO:0019862 | IgA binding(GO:0019862) |
| 4.8 | 23.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 4.0 | 15.9 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 3.6 | 25.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 3.3 | 10.0 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 2.8 | 19.9 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 2.4 | 9.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.8 | 7.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 1.8 | 16.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 1.6 | 4.7 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 1.5 | 6.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 1.4 | 12.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.4 | 5.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 1.3 | 6.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 1.3 | 8.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 1.2 | 16.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.2 | 5.9 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 1.1 | 4.5 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 1.1 | 4.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.0 | 27.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 1.0 | 6.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.0 | 21.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.8 | 8.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.8 | 6.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.7 | 3.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.5 | 2.7 | GO:0033041 | sweet taste receptor activity(GO:0033041) |
| 0.5 | 9.6 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.4 | 51.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 21.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.4 | 5.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 3.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 10.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 5.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 3.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.3 | 0.8 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.2 | 4.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.5 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 188.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 3.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 8.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 1.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 24.9 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.1 | 8.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 7.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 2.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 3.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.3 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 5.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 21.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 4.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 31.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 6.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 7.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 18.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.0 | 9.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 23.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.6 | 10.8 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.6 | 8.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.5 | 6.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 2.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 3.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 5.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 4.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 23.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.6 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 4.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |