Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
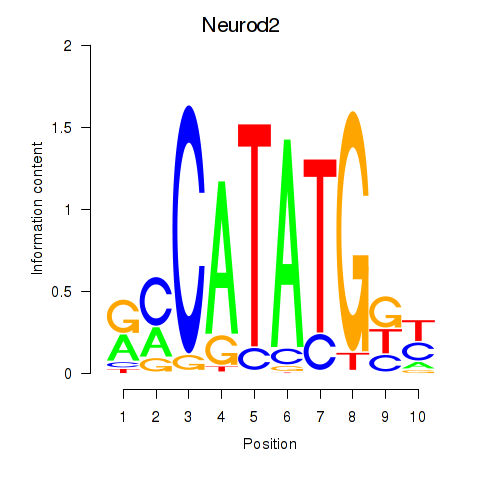
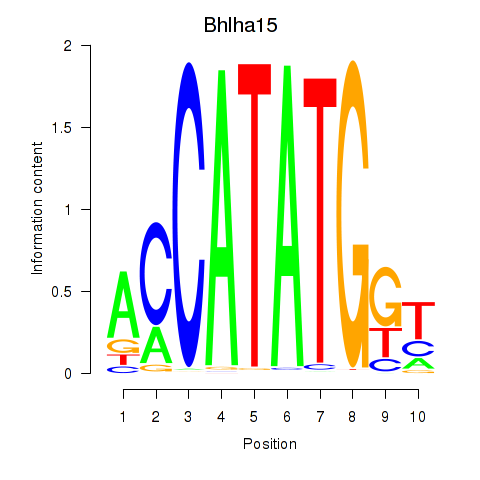
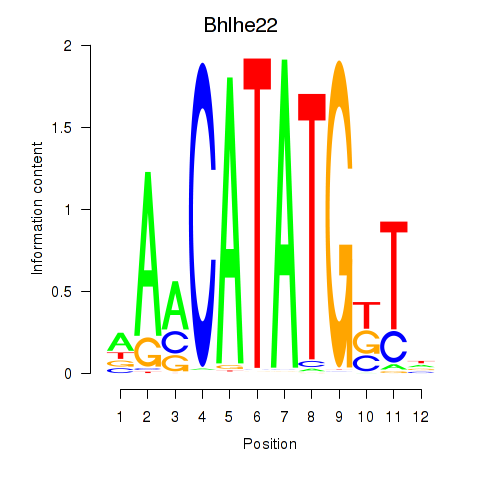
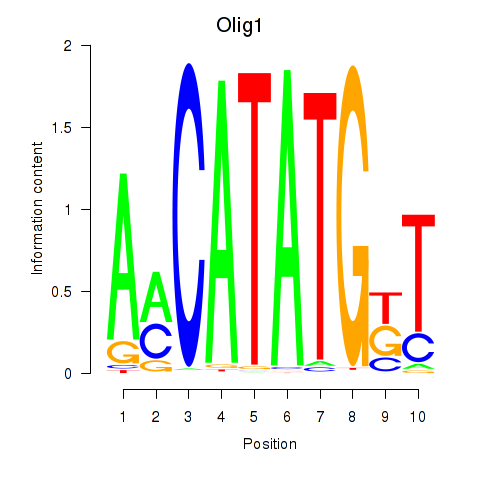
Results for Neurod2_Bhlha15_Bhlhe22_Olig1
Z-value: 0.46
Transcription factors associated with Neurod2_Bhlha15_Bhlhe22_Olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod2
|
ENSRNOG00000028417 | Neurod2 |
|
Bhlha15
|
ENSRNOG00000025164 | basic helix-loop-helix family, member a15 |
|
Bhlhe22
|
ENSRNOG00000021745 | basic helix-loop-helix family, member e22 |
|
Olig1
|
ENSRNOG00000028648 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlha15 | rn6_v1_chr12_-_12407561_12407561 | 0.09 | 1.1e-01 | Click! |
| Bhlhe22 | rn6_v1_chr2_+_102685513_102685513 | 0.08 | 1.8e-01 | Click! |
| Olig1 | rn6_v1_chr11_+_31428358_31428358 | -0.01 | 8.6e-01 | Click! |
Activity profile of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
Sorted Z-values of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_128003418 | 34.41 |
ENSRNOT00000013896
|
Serpina3c
|
serine (or cysteine) proteinase inhibitor, clade A, member 3C |
| chr6_+_128048099 | 28.02 |
ENSRNOT00000084685
ENSRNOT00000087017 |
LOC500712
|
Ab1-233 |
| chr13_+_89597138 | 22.50 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr1_+_148240504 | 21.82 |
ENSRNOT00000085373
|
LOC100361547
|
Cytochrome P450, family 2, subfamily c, polypeptide 7-like |
| chr4_-_31730386 | 18.08 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr7_+_28414350 | 17.11 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr19_-_22632071 | 16.48 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr2_-_100372252 | 13.87 |
ENSRNOT00000011890
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr4_-_82194927 | 12.77 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr15_+_28028521 | 11.87 |
ENSRNOT00000089631
|
AC114343.1
|
|
| chr1_-_189181901 | 9.50 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr17_+_69468427 | 8.11 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr3_-_123179644 | 7.47 |
ENSRNOT00000028835
|
Lzts3
|
leucine zipper tumor suppressor family member 3 |
| chr1_+_189364288 | 7.22 |
ENSRNOT00000080338
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr10_+_99388130 | 6.86 |
ENSRNOT00000006238
|
Kcnj16
|
potassium voltage-gated channel subfamily J member 16 |
| chrX_-_138435391 | 5.66 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr4_+_122365093 | 5.47 |
ENSRNOT00000024011
|
Klf15
|
Kruppel-like factor 15 |
| chr17_+_30556884 | 5.41 |
ENSRNOT00000080929
ENSRNOT00000022022 |
Eci2
|
enoyl-CoA delta isomerase 2 |
| chr8_-_130550388 | 5.31 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr1_-_205630073 | 5.23 |
ENSRNOT00000037064
|
Tex36
|
testis expressed 36 |
| chr8_-_111777602 | 5.14 |
ENSRNOT00000052317
ENSRNOT00000083855 |
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr3_+_128155069 | 5.12 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr6_-_110681408 | 4.77 |
ENSRNOT00000078265
|
Angel1
|
angel homolog 1 |
| chr15_-_109316953 | 4.39 |
ENSRNOT00000019158
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr3_+_40038336 | 4.32 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr10_-_16046033 | 4.30 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_-_16045835 | 4.29 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr5_-_64946580 | 4.17 |
ENSRNOT00000007290
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr11_+_54137639 | 4.12 |
ENSRNOT00000066343
|
LOC100909977
|
leukocyte surface antigen CD47-like |
| chrX_+_6273733 | 3.89 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr6_+_36089433 | 3.86 |
ENSRNOT00000090438
ENSRNOT00000005731 |
Nt5c1b
|
5'-nucleotidase, cytosolic IB |
| chr1_-_201894775 | 3.78 |
ENSRNOT00000050519
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr13_+_56096834 | 3.74 |
ENSRNOT00000035129
|
Dennd1b
|
DENN domain containing 1B |
| chr13_-_56958549 | 3.70 |
ENSRNOT00000017293
ENSRNOT00000083912 |
RGD1564614
|
similar to complement factor H-related protein |
| chr2_+_196334626 | 3.61 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr3_-_66417741 | 3.45 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr19_+_683484 | 3.43 |
ENSRNOT00000090332
|
Terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr4_-_22424862 | 3.29 |
ENSRNOT00000082359
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr17_-_10208360 | 3.24 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr8_+_69971778 | 3.24 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr3_-_39596718 | 3.12 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chrX_-_130794673 | 2.96 |
ENSRNOT00000080612
|
LOC691030
|
similar to RIKEN cDNA 1700001F22 |
| chr3_-_146407252 | 2.85 |
ENSRNOT00000091310
|
Apmap
|
adipocyte plasma membrane associated protein |
| chr7_-_52404774 | 2.79 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr3_-_145810834 | 2.75 |
ENSRNOT00000075429
|
LOC102555217
|
zinc finger protein 120-like |
| chr7_-_132343169 | 2.54 |
ENSRNOT00000021064
|
Abcd2
|
ATP binding cassette subfamily D member 2 |
| chr1_-_266428239 | 2.53 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr17_-_51912496 | 2.51 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr4_-_82280722 | 2.45 |
ENSRNOT00000071668
|
Hoxa9
|
homeobox A9 |
| chr9_-_82477136 | 2.37 |
ENSRNOT00000026753
ENSRNOT00000089555 |
Resp18
|
regulated endocrine-specific protein 18 |
| chr1_+_129613497 | 2.37 |
ENSRNOT00000074410
|
LOC102551255
|
tetraspanin-7-like |
| chr4_+_79868442 | 2.35 |
ENSRNOT00000089973
|
Mpp6
|
membrane palmitoylated protein 6 |
| chr1_-_189238776 | 2.32 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr3_-_78235760 | 2.26 |
ENSRNOT00000008435
|
Olr698
|
olfactory receptor 698 |
| chr3_+_145764932 | 2.23 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr1_+_72810545 | 2.23 |
ENSRNOT00000092117
|
Ptprh
|
protein tyrosine phosphatase, receptor type, H |
| chr5_+_98469047 | 2.12 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr7_-_70328148 | 2.08 |
ENSRNOT00000074645
|
Mettl21b
|
methyltransferase like 21B |
| chr10_-_110308514 | 1.95 |
ENSRNOT00000054927
|
Tex19.2
|
testis expressed gene 19.2 |
| chr10_+_69737328 | 1.84 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr1_+_167719947 | 1.80 |
ENSRNOT00000024971
|
Olr43
|
olfactory receptor 43 |
| chr20_-_1099336 | 1.76 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chr14_+_22107416 | 1.71 |
ENSRNOT00000060179
ENSRNOT00000080114 |
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr2_+_148722231 | 1.71 |
ENSRNOT00000018022
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr10_+_74959285 | 1.67 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr2_+_198721724 | 1.65 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr19_-_42180981 | 1.55 |
ENSRNOT00000019577
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr3_-_38090526 | 1.54 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr13_-_104080631 | 1.50 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr15_-_49505553 | 1.49 |
ENSRNOT00000028974
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr5_-_2982603 | 1.48 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr1_+_205911552 | 1.46 |
ENSRNOT00000024751
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chrY_-_1398030 | 1.43 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr4_-_84740909 | 1.38 |
ENSRNOT00000013088
|
Scrn1
|
secernin 1 |
| chr12_-_38504774 | 1.37 |
ENSRNOT00000011286
|
B3gnt4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr20_+_5535432 | 1.36 |
ENSRNOT00000040859
|
Syngap1
|
synaptic Ras GTPase activating protein 1 |
| chr18_-_19275273 | 1.36 |
ENSRNOT00000041707
|
LOC102547344
|
lateral signaling target protein 2 homolog |
| chr14_+_77922045 | 1.33 |
ENSRNOT00000081198
|
Stk32b
|
serine/threonine kinase 32B |
| chr2_-_259382765 | 1.27 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr18_+_80939875 | 1.26 |
ENSRNOT00000021729
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr2_+_208738132 | 1.23 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chr10_+_36517169 | 1.23 |
ENSRNOT00000040534
|
Olr1408
|
olfactory receptor 1408 |
| chr15_+_27400717 | 1.21 |
ENSRNOT00000047008
|
Olr1620
|
olfactory receptor 1620 |
| chr10_-_12491068 | 1.19 |
ENSRNOT00000071158
|
Olr1368
|
olfactory receptor 1368 |
| chrX_-_111365849 | 1.17 |
ENSRNOT00000088979
|
Nup62cl
|
nucleoporin 62 C-terminal like |
| chr15_-_57651041 | 1.16 |
ENSRNOT00000072138
|
Spert
|
spermatid associated |
| chr10_+_38788992 | 1.16 |
ENSRNOT00000009727
|
Gdf9
|
growth differentiation factor 9 |
| chr13_+_49003803 | 1.15 |
ENSRNOT00000080556
|
Lemd1
|
LEM domain containing 1 |
| chr8_-_43443531 | 1.15 |
ENSRNOT00000007901
|
LOC103693060
|
olfactory receptor 148-like |
| chr3_+_152630415 | 1.15 |
ENSRNOT00000027369
|
Aar2
|
AAR2 splicing factor homolog |
| chr10_-_12731897 | 1.14 |
ENSRNOT00000042582
|
Olr1378
|
olfactory receptor 1378 |
| chr2_+_179952227 | 1.13 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr5_+_104362971 | 1.12 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr10_-_12644292 | 1.11 |
ENSRNOT00000041612
|
Olr1375
|
olfactory receptor 1375 |
| chr4_-_173640684 | 1.10 |
ENSRNOT00000010728
|
Rergl
|
RERG like |
| chr11_+_9642365 | 1.09 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr8_+_19548488 | 1.08 |
ENSRNOT00000008605
|
Olr1148
|
olfactory receptor 1148 |
| chr7_-_145338152 | 1.06 |
ENSRNOT00000055268
|
Spt1
|
salivary protein 1 |
| chr6_+_52663112 | 1.04 |
ENSRNOT00000013842
|
Atxn7l1
|
ataxin 7-like 1 |
| chrX_+_110342718 | 1.02 |
ENSRNOT00000043861
|
AABR07040792.1
|
|
| chr6_+_18880737 | 1.00 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr7_-_145575981 | 0.99 |
ENSRNOT00000074622
|
LOC102555445
|
16.5 kDa submandibular gland glycoprotein-like |
| chr10_-_12370656 | 0.98 |
ENSRNOT00000060995
|
Olr1362
|
olfactory receptor 1362 |
| chrX_-_18341467 | 0.97 |
ENSRNOT00000045802
|
Spin2a
|
spindlin family, member 2A |
| chrX_-_143274180 | 0.97 |
ENSRNOT00000051550
|
Mcf2
|
MCF.2 cell line derived transforming sequence |
| chr20_-_5485837 | 0.95 |
ENSRNOT00000092272
ENSRNOT00000000559 ENSRNOT00000092597 |
Daxx
|
death-domain associated protein |
| chr2_+_54660722 | 0.95 |
ENSRNOT00000060376
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr5_-_66322567 | 0.95 |
ENSRNOT00000032726
|
Cylc2
|
cylicin 2 |
| chr3_-_103460529 | 0.90 |
ENSRNOT00000047274
|
LOC100909573
|
olfactory receptor 4F6-like |
| chr11_+_43194348 | 0.90 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr7_-_118518193 | 0.88 |
ENSRNOT00000075636
|
LOC102555083
|
zinc finger protein 250-like |
| chr19_-_39267928 | 0.87 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chrX_+_122313470 | 0.87 |
ENSRNOT00000018322
|
RGD1566387
|
similar to hypothetical protein |
| chr11_+_55497234 | 0.86 |
ENSRNOT00000070882
|
LOC108348268
|
olfactory receptor 2T33-like |
| chr10_-_91581232 | 0.86 |
ENSRNOT00000066453
ENSRNOT00000068686 |
AABR07030520.1
|
|
| chr3_+_5167685 | 0.85 |
ENSRNOT00000046109
|
Lcn4
|
lipocalin 4 |
| chr10_-_12674077 | 0.85 |
ENSRNOT00000041353
|
Olr1376
|
olfactory receptor 1376 |
| chr5_-_128446725 | 0.84 |
ENSRNOT00000011272
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr16_+_2706428 | 0.83 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr4_-_39102807 | 0.83 |
ENSRNOT00000052063
|
Thsd7a
|
thrombospondin type 1 domain containing 7A |
| chr7_-_120518653 | 0.82 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr2_-_154763094 | 0.79 |
ENSRNOT00000031015
|
Vom2r46
|
vomeronasal 2 receptor, 46 |
| chr4_-_165787668 | 0.79 |
ENSRNOT00000044029
|
Tas2r106
|
taste receptor, type 2, member 106 |
| chr1_+_228690119 | 0.79 |
ENSRNOT00000087169
|
Pfpl
|
pore forming protein-like |
| chr16_+_35573058 | 0.79 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr1_-_8885967 | 0.78 |
ENSRNOT00000016102
|
Gje1
|
gap junction protein, epsilon 1 |
| chr2_+_210381829 | 0.76 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr2_-_2779109 | 0.76 |
ENSRNOT00000017098
|
Rfesd
|
Rieske (Fe-S) domain containing |
| chr9_-_105655471 | 0.76 |
ENSRNOT00000014921
|
RGD1560925
|
similar to 2610034M16Rik protein |
| chr15_+_46179787 | 0.75 |
ENSRNOT00000059213
|
Defb41
|
defensin beta 41 |
| chr16_-_75241303 | 0.75 |
ENSRNOT00000058056
|
Defb2
|
defensin beta 2 |
| chr12_-_18526250 | 0.74 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr9_+_112360419 | 0.73 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr6_-_86223052 | 0.72 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr1_+_89149998 | 0.70 |
ENSRNOT00000056742
|
LOC688924
|
hypothetical protein LOC688924 |
| chr3_+_103637469 | 0.70 |
ENSRNOT00000088881
|
Olr795
|
olfactory receptor 795 |
| chr20_+_649582 | 0.69 |
ENSRNOT00000074145
|
Olr1687
|
olfactory receptor 1687 |
| chr10_-_60006037 | 0.69 |
ENSRNOT00000065541
|
Olr1485
|
olfactory receptor 1485 |
| chr1_+_212651917 | 0.68 |
ENSRNOT00000025635
|
Olr287
|
olfactory receptor 287 |
| chr5_-_12563429 | 0.68 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr3_+_147511563 | 0.68 |
ENSRNOT00000006828
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr8_-_28044876 | 0.67 |
ENSRNOT00000072152
|
Acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr12_+_41600005 | 0.67 |
ENSRNOT00000001872
|
Plbd2
|
phospholipase B domain containing 2 |
| chr12_-_18227991 | 0.67 |
ENSRNOT00000072986
|
AABR07035501.1
|
|
| chr20_-_11626876 | 0.67 |
ENSRNOT00000001635
|
LOC100361664
|
keratin associated protein 12-1-like |
| chrX_-_154918095 | 0.66 |
ENSRNOT00000085224
|
LOC100911991
|
elongation factor 1-alpha 1-like |
| chr4_-_121969446 | 0.66 |
ENSRNOT00000085001
|
Vom1r99
|
vomeronasal 1 receptor 99 |
| chrX_+_156319687 | 0.66 |
ENSRNOT00000091147
ENSRNOT00000079378 |
Fam3a
|
family with sequence similarity 3, member A |
| chr7_+_13378338 | 0.65 |
ENSRNOT00000042747
|
Olr1073
|
olfactory receptor 1073 |
| chr9_+_60900213 | 0.65 |
ENSRNOT00000017859
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr15_+_27438853 | 0.64 |
ENSRNOT00000011636
|
Olr1622
|
olfactory receptor 1622 |
| chr10_-_16910641 | 0.64 |
ENSRNOT00000005273
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr1_+_188278469 | 0.62 |
ENSRNOT00000065601
|
Tmc5
|
transmembrane channel-like 5 |
| chr1_+_164425475 | 0.61 |
ENSRNOT00000023386
|
Klhl35
|
kelch-like family member 35 |
| chr11_+_68633160 | 0.61 |
ENSRNOT00000063795
|
Sec22a
|
SEC22 homolog A, vesicle trafficking protein |
| chr1_-_238381871 | 0.60 |
ENSRNOT00000077601
|
Tmc1
|
transmembrane channel-like 1 |
| chr19_+_24329544 | 0.59 |
ENSRNOT00000080934
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr6_-_127571016 | 0.59 |
ENSRNOT00000057322
|
Serpina1f
|
serine (or cysteine) peptidase inhibitor, clade A, member 1F |
| chr1_+_168655404 | 0.58 |
ENSRNOT00000021307
|
Olr110
|
olfactory receptor 110 |
| chr20_+_29445510 | 0.58 |
ENSRNOT00000044805
|
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr1_-_198706852 | 0.57 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr4_-_67610710 | 0.57 |
ENSRNOT00000039067
|
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr2_-_199529864 | 0.57 |
ENSRNOT00000023742
|
Olr390
|
olfactory receptor 390 |
| chr3_+_138174054 | 0.56 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr6_+_136245027 | 0.56 |
ENSRNOT00000086997
|
LOC103692719
|
tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A |
| chr10_-_60772313 | 0.54 |
ENSRNOT00000050847
|
Olr1504
|
olfactory receptor 1504 |
| chr3_+_147511406 | 0.54 |
ENSRNOT00000082438
|
Slc52a3
|
solute carrier family 52 member 3 |
| chrX_-_153491837 | 0.53 |
ENSRNOT00000089027
|
LOC100360413
|
eukaryotic translation elongation factor 1 alpha 1-like |
| chr20_+_29558689 | 0.52 |
ENSRNOT00000085229
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr5_+_79367663 | 0.52 |
ENSRNOT00000011508
|
Atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr1_+_215135482 | 0.51 |
ENSRNOT00000079062
ENSRNOT00000064138 ENSRNOT00000054866 |
AABR07006042.1
|
|
| chr4_-_130659697 | 0.51 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr19_+_60104804 | 0.50 |
ENSRNOT00000078559
|
Pard3
|
par-3 family cell polarity regulator |
| chr8_-_55171718 | 0.50 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr1_-_217620420 | 0.49 |
ENSRNOT00000092783
|
Cttn
|
cortactin |
| chr7_-_117978787 | 0.48 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr20_-_11644166 | 0.47 |
ENSRNOT00000073009
|
LOC100361739
|
keratin associated protein 12-1-like |
| chr3_+_78876609 | 0.47 |
ENSRNOT00000049936
|
Olr726
|
olfactory receptor 726 |
| chr5_+_141530211 | 0.47 |
ENSRNOT00000056546
|
4933427I04Rik
|
Riken cDNA 4933427I04 gene |
| chr3_+_15788778 | 0.46 |
ENSRNOT00000041240
|
Olr397
|
olfactory receptor 397 |
| chr17_+_87274944 | 0.45 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr3_+_102471249 | 0.45 |
ENSRNOT00000051083
|
Olr749
|
olfactory receptor 749 |
| chrX_+_70354360 | 0.43 |
ENSRNOT00000076852
ENSRNOT00000029046 |
Dgat2l6
|
diacylglycerol O-acyltransferase 2-like 6 |
| chr3_-_173839433 | 0.43 |
ENSRNOT00000075547
|
AC127963.1
|
|
| chr6_-_44363915 | 0.42 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr16_-_75028977 | 0.42 |
ENSRNOT00000058071
|
Defb13
|
defensin beta 13 |
| chr1_+_169074475 | 0.42 |
ENSRNOT00000050248
|
Olr132
|
olfactory receptor 132 |
| chr8_-_43470377 | 0.42 |
ENSRNOT00000050312
|
LOC103690273
|
olfactory receptor 149 |
| chr7_-_120839577 | 0.42 |
ENSRNOT00000018526
|
Dmc1
|
DNA meiotic recombinase 1 |
| chr3_+_37545238 | 0.41 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr10_-_12236107 | 0.41 |
ENSRNOT00000049390
|
Olr1381
|
olfactory receptor 1381 |
| chr2_+_234298839 | 0.41 |
ENSRNOT00000072216
|
Dcm5
|
Dcm5 protein |
| chr3_-_103447716 | 0.39 |
ENSRNOT00000074196
|
Olr791
|
olfactory receptor 791 |
| chr10_-_4910305 | 0.39 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr8_-_43304560 | 0.39 |
ENSRNOT00000060092
|
Olr1307
|
olfactory receptor 1307 |
| chr15_-_28786094 | 0.37 |
ENSRNOT00000044250
|
Olr1639
|
olfactory receptor 1639 |
| chr10_+_34631398 | 0.37 |
ENSRNOT00000046027
|
Olr1391
|
olfactory receptor 1391 |
| chr1_+_168830222 | 0.36 |
ENSRNOT00000050333
|
Olr122
|
olfactory receptor 122 |
| chr1_-_149859479 | 0.36 |
ENSRNOT00000052199
|
LOC100910009
|
olfactory receptor 14A2-like |
| chr5_+_98998163 | 0.35 |
ENSRNOT00000075600
|
Pramef5
|
PRAME family member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod2_Bhlha15_Bhlhe22_Olig1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.5 | GO:0046340 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 4.5 | 18.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 4.3 | 17.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 2.7 | 16.5 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 2.4 | 9.5 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.7 | 5.1 | GO:0035723 | interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) regulation of monocyte extravasation(GO:2000437) |
| 0.9 | 8.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.8 | 2.5 | GO:0010034 | response to acetate(GO:0010034) |
| 0.8 | 3.3 | GO:0043215 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.8 | 21.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.7 | 3.7 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.7 | 33.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.7 | 3.5 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.6 | 3.9 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.5 | 3.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 1.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 1.5 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.4 | 1.1 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.4 | 3.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 1.0 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 0.3 | 2.5 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.3 | 3.4 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.3 | 1.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 3.7 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 | 0.7 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.2 | 1.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 6.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 2.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 5.5 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 5.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 2.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 4.4 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 1.7 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 3.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.6 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) DNA protection(GO:0042262) |
| 0.1 | 0.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 1.5 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 5.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 4.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 1.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 1.3 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.1 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 5.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.1 | 17.7 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.1 | 0.2 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) response to formaldehyde(GO:1904404) |
| 0.1 | 0.5 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.1 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.3 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 1.5 | GO:0072661 | protein targeting to plasma membrane(GO:0072661) |
| 0.0 | 1.4 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.2 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.8 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.3 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 7.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 23.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 1.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 17.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 2.5 | 22.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.8 | 2.5 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.4 | 2.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 3.4 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 0.8 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.3 | 5.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 3.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 4.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 4.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 5.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 9.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.5 | GO:0098871 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.1 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.7 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 6.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.0 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 65.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.5 | GO:0044438 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 7.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 4.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.0 | GO:0044299 | C-fiber(GO:0044299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 5.5 | 16.5 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 3.6 | 18.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.4 | 5.4 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 1.1 | 4.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.9 | 7.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.8 | 17.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.7 | 21.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.7 | 3.3 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.6 | 3.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 2.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.5 | 8.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.4 | 2.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 1.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.4 | 7.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 1.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 3.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 1.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) |
| 0.3 | 35.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 6.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.3 | 2.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 3.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 0.8 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.3 | 1.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 2.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 2.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 5.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) lamin binding(GO:0005521) |
| 0.2 | 3.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 13.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 5.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 7.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 3.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.6 | GO:0047429 | pyrimidine nucleotide binding(GO:0019103) deoxyribonucleotide binding(GO:0032552) nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.7 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.9 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 22.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.7 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 3.9 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 1.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 5.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 22.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.9 | 16.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.6 | 17.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 5.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 18.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 7.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 3.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 2.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 3.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 6.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 6.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 7.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |