Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
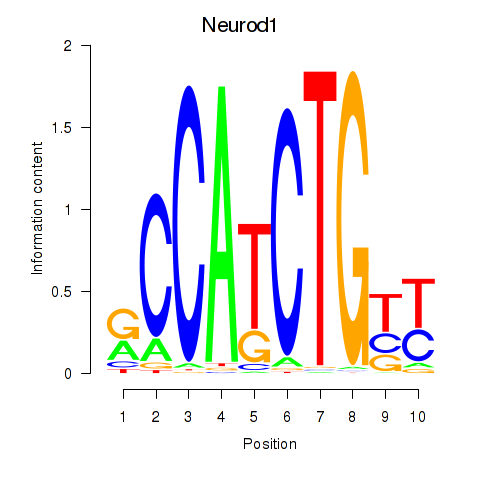
Results for Neurod1
Z-value: 1.29
Transcription factors associated with Neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod1
|
ENSRNOG00000005609 | neuronal differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurod1 | rn6_v1_chr3_-_66417741_66417741 | 0.26 | 1.6e-06 | Click! |
Activity profile of Neurod1 motif
Sorted Z-values of Neurod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_80321585 | 46.62 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr10_+_110631494 | 35.04 |
ENSRNOT00000054915
|
Fn3k
|
fructosamine 3 kinase |
| chr13_+_85818427 | 31.81 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr1_+_101161252 | 28.03 |
ENSRNOT00000028064
ENSRNOT00000064184 |
Slc17a7
|
solute carrier family 17 member 7 |
| chr15_+_34251606 | 26.98 |
ENSRNOT00000025725
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr3_-_2534375 | 25.25 |
ENSRNOT00000037725
|
Grin1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr15_+_43905099 | 24.46 |
ENSRNOT00000016568
|
Ebf2
|
early B-cell factor 2 |
| chr4_-_85915099 | 22.92 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr16_+_46731403 | 22.75 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr4_-_129619142 | 20.67 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr12_+_14021727 | 20.61 |
ENSRNOT00000060608
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chrX_-_15707436 | 20.47 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr17_+_72160735 | 20.43 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr7_-_12429897 | 19.87 |
ENSRNOT00000020670
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr4_+_144382945 | 19.22 |
ENSRNOT00000007601
|
Cav3
|
caveolin 3 |
| chr5_+_145257714 | 19.20 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr7_-_50638798 | 19.15 |
ENSRNOT00000048880
|
Syt1
|
synaptotagmin 1 |
| chr1_+_198655742 | 18.58 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr3_+_113257688 | 18.41 |
ENSRNOT00000019320
|
Map1a
|
microtubule-associated protein 1A |
| chr10_+_34185898 | 16.81 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr15_-_93307420 | 16.09 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr14_-_86047162 | 15.93 |
ENSRNOT00000018227
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr19_+_52032886 | 15.86 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr9_-_32019205 | 15.72 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr14_+_12218553 | 15.33 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr9_-_85243001 | 15.10 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr14_+_60857989 | 14.89 |
ENSRNOT00000034411
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr3_-_72602548 | 14.65 |
ENSRNOT00000031745
|
Lrrc55
|
leucine rich repeat containing 55 |
| chr13_+_52662996 | 14.62 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr17_-_13393243 | 14.57 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr3_-_168018410 | 14.48 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr3_-_38090526 | 14.34 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr14_+_37116492 | 14.21 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr5_-_12563429 | 14.15 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr1_+_263186235 | 14.03 |
ENSRNOT00000021876
|
Cnnm1
|
cyclin and CBS domain divalent metal cation transport mediator 1 |
| chr3_-_2534663 | 13.60 |
ENSRNOT00000049297
ENSRNOT00000044246 |
Grin1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr17_-_84614228 | 13.25 |
ENSRNOT00000043042
|
AABR07028748.1
|
|
| chr19_-_38321528 | 13.23 |
ENSRNOT00000031977
|
Smpd3
|
sphingomyelin phosphodiesterase 3 |
| chr9_-_100253609 | 12.84 |
ENSRNOT00000036061
|
Kif1a
|
kinesin family member 1A |
| chr2_-_184289126 | 12.59 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr10_+_70262361 | 12.52 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr12_-_36398206 | 12.45 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr2_-_171196395 | 12.45 |
ENSRNOT00000013279
|
Bche
|
butyrylcholinesterase |
| chr20_-_27083410 | 11.98 |
ENSRNOT00000000432
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chr7_-_71226150 | 11.57 |
ENSRNOT00000005875
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr15_-_19876389 | 11.30 |
ENSRNOT00000012400
ENSRNOT00000086368 |
Fermt2
|
fermitin family member 2 |
| chr1_-_80783898 | 11.26 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr4_+_180291389 | 11.22 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr4_-_482645 | 11.14 |
ENSRNOT00000062073
ENSRNOT00000071713 |
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr10_-_15928169 | 10.89 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr13_+_80517536 | 10.67 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr16_-_39476384 | 10.39 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr16_-_39476025 | 10.26 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr13_+_52889737 | 10.22 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr4_+_86275717 | 10.17 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr5_-_100647298 | 9.97 |
ENSRNOT00000067538
ENSRNOT00000013092 |
Nfib
|
nuclear factor I/B |
| chr12_-_25638797 | 9.91 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr1_+_80279706 | 9.58 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr9_+_94279155 | 9.50 |
ENSRNOT00000065805
|
Prss56
|
protease, serine, 56 |
| chr19_-_34752695 | 9.49 |
ENSRNOT00000052018
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chrX_+_24891156 | 9.46 |
ENSRNOT00000071173
|
Wwc3
|
WWC family member 3 |
| chr7_-_12274803 | 9.44 |
ENSRNOT00000049146
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr1_+_212651917 | 9.42 |
ENSRNOT00000025635
|
Olr287
|
olfactory receptor 287 |
| chr4_+_113970079 | 9.41 |
ENSRNOT00000079547
|
Rtkn
|
rhotekin |
| chr18_-_57245666 | 9.39 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr4_-_84740909 | 9.35 |
ENSRNOT00000013088
|
Scrn1
|
secernin 1 |
| chr7_+_13378338 | 9.30 |
ENSRNOT00000042747
|
Olr1073
|
olfactory receptor 1073 |
| chr2_-_149088787 | 9.18 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chrX_+_159158194 | 9.18 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr4_+_84854386 | 9.07 |
ENSRNOT00000013620
|
Mturn
|
maturin, neural progenitor differentiation regulator homolog |
| chr10_+_92628356 | 8.97 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr3_+_79713567 | 8.95 |
ENSRNOT00000012110
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr1_+_150310319 | 8.79 |
ENSRNOT00000042081
|
Olr34
|
olfactory receptor 34 |
| chr12_-_46414434 | 8.77 |
ENSRNOT00000041281
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr1_+_145715969 | 8.71 |
ENSRNOT00000037996
|
Tmc3
|
transmembrane channel-like 3 |
| chr2_+_58462949 | 8.67 |
ENSRNOT00000080618
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr2_+_11658568 | 8.57 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr18_+_80939875 | 8.39 |
ENSRNOT00000021729
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr7_+_144595960 | 8.30 |
ENSRNOT00000021909
|
Hoxc9
|
homeobox C9 |
| chr3_+_148386189 | 8.23 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr14_+_36071376 | 8.23 |
ENSRNOT00000082183
|
Lnx1
|
ligand of numb-protein X 1 |
| chr10_-_109729019 | 8.22 |
ENSRNOT00000054959
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr4_+_168752133 | 8.08 |
ENSRNOT00000010289
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr5_-_34813116 | 8.06 |
ENSRNOT00000017479
|
Nkain3
|
Sodium/potassium transporting ATPase interacting 3 |
| chr5_-_100647727 | 7.85 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr8_+_21271190 | 7.78 |
ENSRNOT00000048082
|
LOC100361194
|
olfactory receptor Olr1192-like |
| chr1_+_72420352 | 7.76 |
ENSRNOT00000066307
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr3_-_66417741 | 7.67 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chrX_+_92131209 | 7.62 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr2_+_210381829 | 7.56 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chrX_+_71342775 | 7.49 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr12_-_22021851 | 7.41 |
ENSRNOT00000039280
|
Tsc22d4
|
TSC22 domain family, member 4 |
| chr13_+_92376912 | 7.38 |
ENSRNOT00000050469
|
Olr1600
|
olfactory receptor 1600 |
| chr5_-_40237591 | 7.34 |
ENSRNOT00000011393
|
Fut9
|
fucosyltransferase 9 |
| chr11_+_87374690 | 7.33 |
ENSRNOT00000086117
|
Aifm3
|
apoptosis inducing factor, mitochondria associated 3 |
| chr12_+_12374790 | 7.26 |
ENSRNOT00000001347
|
Tecpr1
|
tectonin beta-propeller repeat containing 1 |
| chr5_-_12526962 | 7.25 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr1_-_70485888 | 7.21 |
ENSRNOT00000020514
|
Olr7
|
olfactory receptor 7 |
| chr5_-_9381214 | 7.15 |
ENSRNOT00000039237
|
RGD1561849
|
similar to RIKEN cDNA 3110035E14 |
| chr2_+_102685513 | 7.00 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr14_-_3300200 | 6.97 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chrX_-_25590048 | 6.93 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr9_+_101268905 | 6.85 |
ENSRNOT00000023270
|
Olr178
|
olfactory receptor 178 |
| chr1_+_70454322 | 6.83 |
ENSRNOT00000072272
|
Olr4
|
olfactory receptor 4 |
| chr9_+_25410669 | 6.83 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr6_-_51018050 | 6.80 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr1_+_162320730 | 6.80 |
ENSRNOT00000035743
|
Kctd21
|
potassium channel tetramerization domain containing 21 |
| chr18_-_28017925 | 6.79 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr6_-_27512287 | 6.76 |
ENSRNOT00000085937
ENSRNOT00000081278 ENSRNOT00000081323 |
Selenoi
|
selenoprotein I |
| chr10_+_65772443 | 6.75 |
ENSRNOT00000013296
|
Sebox
|
SEBOX homeobox |
| chr2_-_172361779 | 6.65 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr7_-_63578490 | 6.62 |
ENSRNOT00000007295
|
Rassf3
|
Ras association domain family member 3 |
| chr4_-_158704170 | 6.53 |
ENSRNOT00000082009
|
Ntf3
|
neurotrophin 3 |
| chr1_+_167870452 | 6.52 |
ENSRNOT00000025027
|
Olr63
|
olfactory receptor 63 |
| chr9_+_99795678 | 6.48 |
ENSRNOT00000056601
|
Olr1353
|
olfactory receptor 1353 |
| chr7_+_71065197 | 6.47 |
ENSRNOT00000051075
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr7_+_70980422 | 6.45 |
ENSRNOT00000077912
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr17_+_87274944 | 6.41 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr7_-_9592000 | 6.39 |
ENSRNOT00000011188
|
Olr1071
|
olfactory receptor 1071 |
| chr1_+_70480941 | 6.33 |
ENSRNOT00000072404
|
Olr6
|
olfactory receptor 6 |
| chr7_+_6644643 | 6.25 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr13_+_60435946 | 6.23 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr4_-_125929002 | 6.19 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr15_-_86105273 | 6.11 |
ENSRNOT00000012600
ENSRNOT00000064942 |
Tbc1d4
|
TBC1 domain family, member 4 |
| chr3_-_81304181 | 6.08 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr4_+_145238947 | 6.08 |
ENSRNOT00000067396
ENSRNOT00000091934 |
Cpne9
|
copine family member 9 |
| chr1_+_82419947 | 5.98 |
ENSRNOT00000027964
|
B3gnt8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr1_-_125967756 | 5.95 |
ENSRNOT00000031488
|
Fam189a1
|
family with sequence similarity 189, member A1 |
| chr1_-_70496347 | 5.87 |
ENSRNOT00000050607
|
Olr8
|
olfactory receptor 8 |
| chr3_+_16183322 | 5.82 |
ENSRNOT00000072630
|
Olr406
|
olfactory receptor 406 |
| chr8_+_68526093 | 5.63 |
ENSRNOT00000011385
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr1_-_228226354 | 5.57 |
ENSRNOT00000029224
|
Olr318
|
olfactory receptor 318 |
| chr3_-_110556808 | 5.51 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr10_-_89816491 | 5.45 |
ENSRNOT00000028220
|
Meox1
|
mesenchyme homeobox 1 |
| chr8_-_21341431 | 5.40 |
ENSRNOT00000044249
|
Olr1192
|
olfactory receptor 1192 |
| chr4_+_72566790 | 5.31 |
ENSRNOT00000007200
|
Olr816
|
olfactory receptor 816 |
| chr8_+_70112925 | 5.30 |
ENSRNOT00000082401
|
Megf11
|
multiple EGF-like-domains 11 |
| chr4_+_155654911 | 5.30 |
ENSRNOT00000087883
|
Foxj2
|
forkhead box J2 |
| chr1_+_213334956 | 5.29 |
ENSRNOT00000073278
|
Olr309
|
olfactory receptor 309 |
| chr10_-_44175030 | 5.25 |
ENSRNOT00000047974
|
LOC103690286
|
olfactory receptor 2T29-like |
| chr3_+_37545238 | 5.16 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr7_-_13531526 | 5.09 |
ENSRNOT00000048054
|
Olr1081
|
olfactory receptor 1081 |
| chr10_+_60974225 | 5.05 |
ENSRNOT00000042349
|
Olr1511
|
olfactory receptor 1511 |
| chr7_+_71057911 | 5.04 |
ENSRNOT00000037218
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr1_+_84411726 | 5.01 |
ENSRNOT00000025303
|
Akt2
|
AKT serine/threonine kinase 2 |
| chr16_-_40025401 | 4.99 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr20_-_11626876 | 4.97 |
ENSRNOT00000001635
|
LOC100361664
|
keratin associated protein 12-1-like |
| chr1_+_163445527 | 4.96 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr5_-_137874692 | 4.95 |
ENSRNOT00000089549
|
Olr867
|
olfactory receptor 867 |
| chr2_-_143104412 | 4.93 |
ENSRNOT00000058116
|
Ufm1
|
ubiquitin-fold modifier 1 |
| chr8_+_43606676 | 4.92 |
ENSRNOT00000083262
|
Olr1319
|
olfactory receptor 1319 |
| chr7_+_9341613 | 4.89 |
ENSRNOT00000044955
|
AABR07055760.1
|
|
| chr7_-_6803318 | 4.89 |
ENSRNOT00000084203
|
Olr954
|
olfactory receptor 954 |
| chr10_-_60772313 | 4.83 |
ENSRNOT00000050847
|
Olr1504
|
olfactory receptor 1504 |
| chr3_+_159902441 | 4.82 |
ENSRNOT00000089893
ENSRNOT00000011978 |
Hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr13_-_90443157 | 4.82 |
ENSRNOT00000006862
|
Nhlh1
|
nescient helix loop helix 1 |
| chr10_+_17542374 | 4.80 |
ENSRNOT00000064079
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr1_+_71434819 | 4.78 |
ENSRNOT00000059069
|
Zfp787
|
zinc finger protein 787 |
| chr15_+_110114148 | 4.73 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr11_-_86303453 | 4.72 |
ENSRNOT00000071453
|
LOC498122
|
similar to CG15908-PA |
| chr4_+_157125998 | 4.72 |
ENSRNOT00000078349
|
C1r
|
complement C1r |
| chr1_-_172322795 | 4.72 |
ENSRNOT00000075318
|
RGD1562400
|
similar to olfactory receptor MOR204-14 |
| chr10_-_44082313 | 4.69 |
ENSRNOT00000051126
|
Olr1421
|
olfactory receptor 1421 |
| chr10_-_44122978 | 4.63 |
ENSRNOT00000045318
|
Olr1423
|
olfactory receptor 1423 |
| chr16_-_20873344 | 4.59 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr7_-_14117687 | 4.56 |
ENSRNOT00000012907
|
Olr1088
|
olfactory receptor 1088 |
| chr2_+_46186105 | 4.56 |
ENSRNOT00000071256
|
LOC100910378
|
olfactory receptor 145-like |
| chr9_+_51009116 | 4.53 |
ENSRNOT00000039313
|
Mettl21cl1
|
methyltransferase like 21C-like 1 |
| chr15_+_27330558 | 4.51 |
ENSRNOT00000085155
|
LOC103693383
|
olfactory receptor 11G2-like |
| chr3_+_15895191 | 4.49 |
ENSRNOT00000047879
|
Olr400
|
olfactory receptor 400 |
| chr7_+_5593735 | 4.44 |
ENSRNOT00000042179
|
Olr917
|
olfactory receptor 917 |
| chr7_-_6626285 | 4.40 |
ENSRNOT00000074329
|
Olr964
|
olfactory receptor 964 |
| chr5_+_18901039 | 4.40 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr3_-_77095635 | 4.34 |
ENSRNOT00000051966
|
Olr650
|
olfactory receptor 650 |
| chr10_+_59173268 | 4.33 |
ENSRNOT00000013486
|
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr1_+_169645330 | 4.33 |
ENSRNOT00000075036
|
Olr159
|
olfactory receptor 159 |
| chr7_-_9485930 | 4.33 |
ENSRNOT00000011198
|
Olr1070
|
olfactory receptor 1070 |
| chr1_+_172157739 | 4.27 |
ENSRNOT00000044619
|
Olr242
|
olfactory receptor 242 |
| chr4_+_153217782 | 4.18 |
ENSRNOT00000015499
|
Cecr2
|
CECR2, histone acetyl-lysine reader |
| chr4_+_117679342 | 4.18 |
ENSRNOT00000021272
|
Figla
|
folliculogenesis specific bHLH transcription factor |
| chr20_+_1944801 | 4.17 |
ENSRNOT00000075060
|
Olr1750
|
olfactory receptor 1750 |
| chrX_-_10218583 | 4.17 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr3_+_15823220 | 4.15 |
ENSRNOT00000075496
|
Olr398
|
olfactory receptor 398 |
| chr12_-_46920952 | 4.14 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr8_-_55171718 | 4.14 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr1_-_228263198 | 4.13 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr12_-_13998172 | 4.09 |
ENSRNOT00000001476
|
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr1_+_169660076 | 4.09 |
ENSRNOT00000073447
|
Olr160
|
olfactory receptor 160 |
| chr2_+_187347602 | 4.08 |
ENSRNOT00000025384
|
Nes
|
nestin |
| chr1_+_169552487 | 4.07 |
ENSRNOT00000023119
|
Olr152
|
olfactory receptor 152 |
| chr2_-_211235440 | 4.06 |
ENSRNOT00000033015
ENSRNOT00000091896 |
Sars
|
seryl-tRNA synthetase |
| chr1_+_172792874 | 4.04 |
ENSRNOT00000077837
|
Olr270
|
olfactory receptor 270 |
| chr8_+_43722228 | 4.04 |
ENSRNOT00000084016
|
Olr1325
|
olfactory receptor 1325 |
| chr1_-_77721737 | 4.02 |
ENSRNOT00000058263
|
LOC103691042
|
transcription elongation factor B polypeptide 2 pseudogene |
| chrX_+_156655960 | 4.02 |
ENSRNOT00000085723
|
Mecp2
|
methyl CpG binding protein 2 |
| chr1_-_149787308 | 4.00 |
ENSRNOT00000088514
|
LOC100909966
|
olfactory receptor 6F1-like |
| chr18_-_59819113 | 4.00 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr3_+_21208600 | 3.98 |
ENSRNOT00000047764
|
Olr427
|
olfactory receptor 427 |
| chr1_+_213194827 | 3.98 |
ENSRNOT00000044878
|
Olr306
|
olfactory receptor 306 |
| chr15_-_36213666 | 3.92 |
ENSRNOT00000078704
|
Olr1291
|
olfactory receptor 1291 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.9 | 38.8 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 7.0 | 28.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 6.8 | 20.5 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 5.5 | 21.8 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 5.3 | 15.9 | GO:0036115 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 5.1 | 15.3 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 4.9 | 14.6 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 4.8 | 19.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 4.2 | 12.6 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 4.1 | 12.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 4.1 | 8.2 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 3.9 | 27.0 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 3.2 | 9.5 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 3.1 | 15.7 | GO:0061743 | motor learning(GO:0061743) |
| 2.8 | 19.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 2.7 | 10.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) ERBB3 signaling pathway(GO:0038129) |
| 2.6 | 20.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 2.2 | 18.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 2.2 | 13.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 2.1 | 8.6 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 2.1 | 22.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.9 | 24.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 1.7 | 8.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.7 | 5.0 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 1.6 | 6.5 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 1.6 | 9.5 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.5 | 7.7 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.5 | 7.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 1.5 | 10.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.4 | 14.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 1.4 | 19.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.2 | 6.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 1.2 | 4.8 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 1.2 | 6.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.2 | 3.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.1 | 22.9 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 1.1 | 14.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.1 | 5.5 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 1.1 | 31.8 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 1.0 | 9.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 1.0 | 4.1 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 1.0 | 6.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 1.0 | 15.1 | GO:0050930 | eosinophil chemotaxis(GO:0048245) induction of positive chemotaxis(GO:0050930) |
| 0.9 | 0.9 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.9 | 14.6 | GO:1900745 | activation of MAPKKK activity(GO:0000185) positive regulation of p38MAPK cascade(GO:1900745) |
| 0.9 | 35.0 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.8 | 6.8 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.8 | 3.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.7 | 2.2 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.7 | 42.4 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.7 | 19.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.7 | 12.0 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.6 | 21.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.6 | 20.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.6 | 25.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.6 | 6.8 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.6 | 3.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.6 | 9.6 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.6 | 1.1 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.6 | 6.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 11.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.5 | 20.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.5 | 2.6 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.5 | 5.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.5 | 6.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.5 | 4.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.4 | 6.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.4 | 1.8 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.4 | 2.2 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.4 | 5.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 8.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 507.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.4 | 7.0 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.4 | 3.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 11.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 3.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 9.0 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.3 | 6.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.3 | 3.7 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.3 | 2.5 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.3 | 8.4 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.3 | 2.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 1.5 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 7.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.2 | 4.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.2 | 3.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 9.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 5.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 12.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 0.9 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.2 | 2.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 | 12.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 18.4 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 0.8 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.5 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 1.7 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 4.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 3.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 7.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 4.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 18.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.1 | 3.0 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 12.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 4.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 3.9 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 11.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 8.8 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 4.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 12.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 4.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.7 | GO:0006914 | autophagy(GO:0006914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 38.8 | GO:0044307 | dendritic branch(GO:0044307) |
| 4.6 | 45.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 3.7 | 14.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 3.6 | 10.7 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 3.2 | 19.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 2.5 | 12.6 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 2.5 | 19.9 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 2.4 | 45.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 2.0 | 6.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.9 | 13.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 1.3 | 15.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.3 | 19.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.2 | 12.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.1 | 15.1 | GO:0031045 | dense core granule(GO:0031045) |
| 1.0 | 20.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.9 | 3.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 11.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.8 | 15.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.7 | 9.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 33.5 | GO:0031672 | A band(GO:0031672) |
| 0.6 | 1.8 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.5 | 2.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 1.5 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.5 | 20.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 5.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.4 | 4.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.4 | 14.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.4 | 9.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 14.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 4.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 7.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 2.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 4.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 4.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.3 | 9.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 18.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 10.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 18.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 9.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 12.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 8.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 1.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 18.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 9.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 6.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 16.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 4.1 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 3.6 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 7.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 3.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 26.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 4.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 6.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 379.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 2.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 5.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 6.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 38.8 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 6.7 | 46.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.6 | 28.0 | GO:0015321 | inorganic phosphate transmembrane transporter activity(GO:0005315) sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 5.3 | 31.8 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 4.1 | 12.4 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 4.0 | 15.9 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 3.8 | 19.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 3.7 | 14.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 3.2 | 18.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 3.1 | 15.3 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 2.5 | 12.6 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 2.4 | 19.2 | GO:0071253 | connexin binding(GO:0071253) |
| 2.3 | 6.8 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 2.1 | 8.4 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) |
| 1.9 | 13.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.6 | 24.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.3 | 10.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.2 | 6.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.2 | 3.7 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 1.2 | 3.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 7.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.2 | 6.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 1.2 | 9.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.0 | 6.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 1.0 | 19.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 8.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 2.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.8 | 6.5 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.8 | 18.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.8 | 4.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.8 | 14.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 3.0 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.7 | 24.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.7 | 20.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 3.4 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.7 | 9.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.6 | 3.2 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.6 | 15.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.5 | 3.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.5 | 19.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 6.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 15.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.4 | 0.8 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.4 | 4.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 18.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 4.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.4 | 507.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.4 | 2.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 8.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.3 | 12.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 7.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 4.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 7.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.3 | 1.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 16.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 2.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 22.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 4.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 7.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 3.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 12.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 7.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 3.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 8.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 2.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 32.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 9.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 3.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 16.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 4.1 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 8.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 6.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 8.2 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 6.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 4.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 3.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.6 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 2.4 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 12.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 19.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.7 | 23.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.6 | 25.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 14.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.5 | 15.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.5 | 14.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 12.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 17.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 13.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 9.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 8.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 10.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 8.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.8 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 5.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 4.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 38.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 2.1 | 47.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.7 | 15.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.2 | 19.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.9 | 12.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.8 | 17.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.7 | 3.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.6 | 23.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 41.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.4 | 8.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.4 | 15.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 12.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 4.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 14.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 13.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 2.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 44.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 5.0 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.2 | 4.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 6.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |