Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Nanog
Z-value: 0.89
Transcription factors associated with Nanog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nanog
|
ENSRNOG00000008368 | Nanog homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nanog | rn6_v1_chr4_+_155531906_155531906 | -0.25 | 6.8e-06 | Click! |
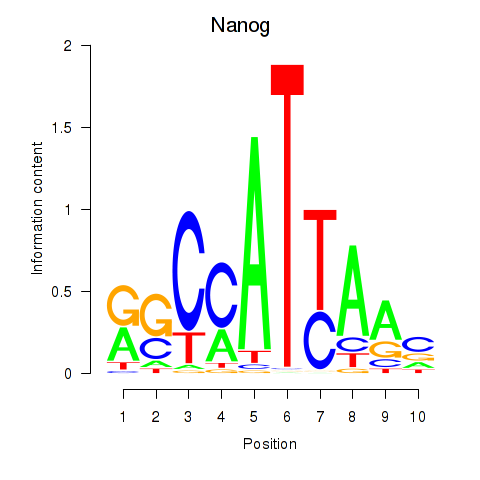
Activity profile of Nanog motif
Sorted Z-values of Nanog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_9762813 | 32.00 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr3_+_117421604 | 22.23 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr13_+_44424689 | 20.64 |
ENSRNOT00000005206
|
Acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr5_+_33784715 | 18.18 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr10_+_49231730 | 17.00 |
ENSRNOT00000065335
|
Trim16
|
tripartite motif-containing 16 |
| chr17_+_69468427 | 16.45 |
ENSRNOT00000058413
|
RGD1564865
|
similar to 20-alpha-hydroxysteroid dehydrogenase |
| chr1_-_224533219 | 14.80 |
ENSRNOT00000051289
|
Ust5r
|
integral membrane transport protein UST5r |
| chr7_+_29435444 | 14.51 |
ENSRNOT00000008613
|
Slc5a8
|
solute carrier family 5 member 8 |
| chr6_+_76349362 | 14.01 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr20_+_26999795 | 13.61 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr9_-_26707571 | 12.13 |
ENSRNOT00000080948
|
AABR07067023.1
|
|
| chr10_+_75620470 | 11.81 |
ENSRNOT00000013882
|
Ccdc182
|
coiled-coil domain containing 182 |
| chr16_+_54319377 | 11.35 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr19_-_10450186 | 11.22 |
ENSRNOT00000088048
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr18_+_29352749 | 11.05 |
ENSRNOT00000025137
|
Slc4a9
|
solute carrier family 4 member 9 |
| chr13_+_55274199 | 10.86 |
ENSRNOT00000029679
|
Atp6v1g3
|
ATPase H+ transporting V1 subunit G3 |
| chr18_-_69944632 | 9.93 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr17_-_78735324 | 9.91 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_-_216663720 | 9.64 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr16_-_48981980 | 9.61 |
ENSRNOT00000014235
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr1_+_177569618 | 9.57 |
ENSRNOT00000090042
|
Tead1
|
TEA domain transcription factor 1 |
| chr20_+_20236151 | 9.26 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr10_-_34333305 | 9.26 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr13_+_99335020 | 9.05 |
ENSRNOT00000029787
|
AABR07021930.1
|
|
| chr15_+_51065316 | 8.95 |
ENSRNOT00000020753
|
Nkx3-1
|
NK3 homeobox 1 |
| chr17_-_42031265 | 8.75 |
ENSRNOT00000068021
|
Dcdc2
|
doublecortin domain containing 2 |
| chr18_-_26656879 | 8.62 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chrX_-_142164220 | 8.51 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr1_-_189238776 | 8.19 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr12_-_44381289 | 8.04 |
ENSRNOT00000001493
|
Nos1
|
nitric oxide synthase 1 |
| chr14_-_19072677 | 8.01 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr16_-_31972507 | 7.90 |
ENSRNOT00000032199
|
Cbr4
|
carbonyl reductase 4 |
| chr7_-_50638798 | 7.81 |
ENSRNOT00000048880
|
Syt1
|
synaptotagmin 1 |
| chr10_+_59799123 | 7.71 |
ENSRNOT00000026493
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr15_-_8989580 | 7.65 |
ENSRNOT00000061402
|
Thrb
|
thyroid hormone receptor beta |
| chr8_-_40883880 | 7.59 |
ENSRNOT00000075593
|
LOC102550797
|
disks large homolog 5-like |
| chr3_-_76496283 | 7.59 |
ENSRNOT00000044305
|
Olr611
|
olfactory receptor 611 |
| chr8_-_78233430 | 7.50 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chrM_+_11736 | 7.38 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr9_+_43259709 | 7.23 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr8_-_128711221 | 7.18 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr16_+_2706428 | 7.11 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr13_-_48927483 | 7.07 |
ENSRNOT00000010976
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr5_+_120340646 | 6.90 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr8_-_115145810 | 6.86 |
ENSRNOT00000079470
|
Abhd14a
|
abhydrolase domain containing 14A |
| chr10_+_85628491 | 6.85 |
ENSRNOT00000085000
ENSRNOT00000066899 |
Cisd3
|
CDGSH iron sulfur domain 3 |
| chr14_+_77922045 | 6.84 |
ENSRNOT00000081198
|
Stk32b
|
serine/threonine kinase 32B |
| chr9_-_82336806 | 6.58 |
ENSRNOT00000024667
|
Slc23a3
|
solute carrier family 23, member 3 |
| chr4_-_22424862 | 6.50 |
ENSRNOT00000082359
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr4_-_82194927 | 6.49 |
ENSRNOT00000072302
|
LOC103692128
|
homeobox protein Hox-A9 |
| chr4_+_1355820 | 6.48 |
ENSRNOT00000072554
|
LOC100911438
|
olfactory receptor 143-like |
| chr10_-_91661558 | 6.47 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr15_-_105402708 | 6.35 |
ENSRNOT00000057564
|
Oxgr1
|
oxoglutarate receptor 1 |
| chr4_+_174181644 | 6.29 |
ENSRNOT00000011555
|
Capza3
|
capping actin protein of muscle Z-line alpha subunit 3 |
| chr11_-_60251415 | 6.16 |
ENSRNOT00000080148
ENSRNOT00000086602 |
Slc9c1
|
solute carrier family 9 member C1 |
| chr14_+_71542057 | 6.13 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr4_+_68849033 | 6.12 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr20_+_10123651 | 6.09 |
ENSRNOT00000001559
|
Pde9a
|
phosphodiesterase 9A |
| chr16_-_15798974 | 6.04 |
ENSRNOT00000046842
ENSRNOT00000065946 |
Nrg3
|
neuregulin 3 |
| chr7_-_134722215 | 6.03 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chr1_+_65851060 | 5.99 |
ENSRNOT00000036880
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr4_+_78168117 | 5.97 |
ENSRNOT00000010853
|
Atp6v0e2
|
ATPase, H+ transporting V0 subunit e2 |
| chr10_+_63677396 | 5.85 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chr7_-_3386522 | 5.78 |
ENSRNOT00000010760
|
Mettl7b
|
methyltransferase like 7B |
| chr5_+_137546860 | 5.75 |
ENSRNOT00000074431
|
Olr858
|
olfactory receptor 858 |
| chrM_+_5323 | 5.62 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr1_+_213511874 | 5.54 |
ENSRNOT00000078080
ENSRNOT00000016883 |
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1 |
| chr1_+_215610368 | 5.43 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chr1_+_80973739 | 5.34 |
ENSRNOT00000026088
ENSRNOT00000081983 |
AC118165.1
|
|
| chr17_+_43460782 | 5.30 |
ENSRNOT00000059494
|
Slc17a4
|
solute carrier family 17, member 4 |
| chr18_+_81694808 | 5.26 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr8_+_42001912 | 5.23 |
ENSRNOT00000071709
|
Olr1226
|
olfactory receptor 1226 |
| chr9_-_105693357 | 5.18 |
ENSRNOT00000066968
|
Nudt12
|
nudix hydrolase 12 |
| chrX_+_13992064 | 5.17 |
ENSRNOT00000036543
|
LOC100363125
|
rCG42854-like |
| chr16_-_47535358 | 5.13 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr10_-_89005213 | 5.08 |
ENSRNOT00000027159
|
Psmc3ip
|
PSMC3 interacting protein |
| chr2_-_231409988 | 5.03 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr10_-_65692016 | 5.00 |
ENSRNOT00000085074
ENSRNOT00000038690 |
Slc13a2
|
solute carrier family 13 member 2 |
| chr18_+_30592794 | 5.00 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chrX_-_152110010 | 4.95 |
ENSRNOT00000044666
|
LOC100362263
|
melanoma antigen family A, 5-like |
| chr10_+_53466870 | 4.93 |
ENSRNOT00000057503
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr3_+_147511406 | 4.92 |
ENSRNOT00000082438
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr20_+_4178686 | 4.87 |
ENSRNOT00000080641
|
Tesb
|
testis specific basic protein |
| chr7_+_60015998 | 4.86 |
ENSRNOT00000007309
|
Best3
|
bestrophin 3 |
| chr11_-_27971359 | 4.83 |
ENSRNOT00000085629
ENSRNOT00000051060 ENSRNOT00000042581 ENSRNOT00000050073 ENSRNOT00000081066 |
Grik1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr1_-_52495582 | 4.78 |
ENSRNOT00000067142
|
Pde10a
|
phosphodiesterase 10A |
| chr10_+_34402482 | 4.78 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr7_-_117289961 | 4.73 |
ENSRNOT00000042642
|
Plec
|
plectin |
| chr6_-_92121351 | 4.69 |
ENSRNOT00000006374
|
Cdkl1
|
cyclin dependent kinase like 1 |
| chr7_+_77066955 | 4.68 |
ENSRNOT00000008700
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr8_-_43854765 | 4.68 |
ENSRNOT00000081506
|
Olr1335
|
olfactory receptor 1335 |
| chr3_+_73327828 | 4.65 |
ENSRNOT00000071299
|
LOC684683
|
similar to olfactory receptor Olr470 |
| chr8_+_39687269 | 4.53 |
ENSRNOT00000012060
|
Tmem218
|
transmembrane protein 218 |
| chr10_+_36466229 | 4.49 |
ENSRNOT00000004865
|
Olr1404
|
olfactory receptor 1404 |
| chr1_+_60717386 | 4.49 |
ENSRNOT00000015019
|
Ppp2r1a
|
protein phosphatase 2 scaffold subunit A alpha |
| chr19_-_24732024 | 4.48 |
ENSRNOT00000037608
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr5_-_159119171 | 4.43 |
ENSRNOT00000074074
ENSRNOT00000075382 ENSRNOT00000074378 |
Arhgef10l
|
Rho guanine nucleotide exchange factor 10 like |
| chr3_+_75906945 | 4.42 |
ENSRNOT00000047110
|
Olr586
|
olfactory receptor 586 |
| chr1_+_247869756 | 4.42 |
ENSRNOT00000054764
|
Mlana
|
melan-A |
| chr2_-_231409496 | 4.38 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chrX_-_65335987 | 4.38 |
ENSRNOT00000047128
|
AABR07038981.1
|
|
| chr17_+_56935451 | 4.37 |
ENSRNOT00000058966
|
RGD1564129
|
similar to hypothetical protein 4930474N05 |
| chrX_+_77065397 | 4.30 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr1_+_168575090 | 4.26 |
ENSRNOT00000048299
|
Olr103
|
olfactory receptor 103 |
| chr3_+_147511563 | 4.24 |
ENSRNOT00000006828
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr16_+_54332660 | 4.20 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr1_-_7322424 | 4.16 |
ENSRNOT00000066725
ENSRNOT00000081112 |
Zc2hc1b
|
zinc finger, C2HC-type containing 1B |
| chr4_-_727691 | 4.14 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr8_+_55037750 | 4.11 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr10_+_45297937 | 4.09 |
ENSRNOT00000066367
|
Trim17
|
tripartite motif-containing 17 |
| chr5_-_136098013 | 4.09 |
ENSRNOT00000089166
|
RGD1563714
|
RGD1563714 |
| chr5_-_17399801 | 4.07 |
ENSRNOT00000011900
|
RGD1563405
|
similar to protein tyrosine phosphatase 4a2 |
| chr1_-_169321075 | 4.06 |
ENSRNOT00000055216
|
RGD1562433
|
similar to ubiquilin 1 isoform 2 |
| chr9_-_74048244 | 4.06 |
ENSRNOT00000018293
|
Lancl1
|
LanC like 1 |
| chr10_-_87248572 | 4.06 |
ENSRNOT00000066637
ENSRNOT00000085677 |
Krt26
|
keratin 26 |
| chr9_-_98551410 | 4.05 |
ENSRNOT00000066346
ENSRNOT00000086678 |
Hes6
|
hes family bHLH transcription factor 6 |
| chr11_+_76147205 | 4.01 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr1_-_206394346 | 3.98 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chrX_+_33884499 | 3.98 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr15_-_52320385 | 3.96 |
ENSRNOT00000067776
|
Dmtn
|
dematin actin binding protein |
| chr2_-_250805445 | 3.94 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr2_-_181874223 | 3.93 |
ENSRNOT00000035846
|
Rbm46
|
RNA binding motif protein 46 |
| chr7_+_3630950 | 3.90 |
ENSRNOT00000074535
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr1_+_258074860 | 3.89 |
ENSRNOT00000054729
|
Cyp2c24
|
cytochrome P450, family 2, subfamily c, polypeptide 24 |
| chr3_-_9738752 | 3.85 |
ENSRNOT00000045993
|
Ptges
|
prostaglandin E synthase |
| chr20_+_31892515 | 3.85 |
ENSRNOT00000071394
|
Tacr2
|
tachykinin receptor 2 |
| chr4_-_176909075 | 3.84 |
ENSRNOT00000067489
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr6_+_8219385 | 3.84 |
ENSRNOT00000040509
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr1_+_212651917 | 3.83 |
ENSRNOT00000025635
|
Olr287
|
olfactory receptor 287 |
| chr13_+_71305548 | 3.83 |
ENSRNOT00000074864
|
LOC684709
|
similar to putative membrane protein Re9 |
| chr8_+_62779875 | 3.82 |
ENSRNOT00000010831
|
Cyp11a1
|
cytochrome P450, family 11, subfamily a, polypeptide 1 |
| chr15_-_9086282 | 3.82 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr5_+_156026911 | 3.80 |
ENSRNOT00000046802
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr5_-_101166651 | 3.77 |
ENSRNOT00000078862
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr10_-_60275762 | 3.77 |
ENSRNOT00000074380
|
Olr1498
|
olfactory receptor 1498 |
| chr1_+_67025240 | 3.75 |
ENSRNOT00000051024
|
Vom1r47
|
vomeronasal 1 receptor 47 |
| chrX_-_23139694 | 3.74 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr7_+_63747577 | 3.65 |
ENSRNOT00000086123
ENSRNOT00000067395 ENSRNOT00000080052 |
RGD1561648
|
RGD1561648 |
| chr7_-_139929895 | 3.65 |
ENSRNOT00000047272
|
Olr1104
|
olfactory receptor 1104 |
| chr17_+_90696019 | 3.64 |
ENSRNOT00000003438
|
Gpr137b
|
G protein-coupled receptor 137B |
| chr14_+_34389991 | 3.63 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr3_-_77227036 | 3.61 |
ENSRNOT00000072638
|
LOC103691843
|
olfactory receptor 4P4-like |
| chr10_+_36517169 | 3.59 |
ENSRNOT00000040534
|
Olr1408
|
olfactory receptor 1408 |
| chr8_-_55194692 | 3.57 |
ENSRNOT00000068366
|
RGD1564937
|
similar to RIKEN cDNA 1110032A03 |
| chrX_+_121612952 | 3.56 |
ENSRNOT00000022122
|
AABR07041179.1
|
|
| chr3_-_77826506 | 3.46 |
ENSRNOT00000071385
|
Olr675
|
olfactory receptor 675 |
| chr5_-_137617258 | 3.46 |
ENSRNOT00000071641
|
Olfr1330-ps1
|
olfactory receptor 1330, pseudogene 1 |
| chrM_+_9451 | 3.43 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr10_+_36485605 | 3.42 |
ENSRNOT00000048759
|
Olr1406
|
olfactory receptor 1406 |
| chr7_-_130120579 | 3.42 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr17_-_84830185 | 3.42 |
ENSRNOT00000040697
|
Skida1
|
SKI/DACH domain containing 1 |
| chrX_+_110789269 | 3.35 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_257970345 | 3.33 |
ENSRNOT00000088853
|
Cyp2c11
|
cytochrome P450, subfamily 2, polypeptide 11 |
| chr3_+_75712483 | 3.31 |
ENSRNOT00000049264
|
Olr575
|
olfactory receptor 575 |
| chr3_-_44501115 | 3.29 |
ENSRNOT00000087071
|
Acvr1
|
activin A receptor type 1 |
| chr1_-_67206713 | 3.22 |
ENSRNOT00000048195
|
Vom1r43
|
vomeronasal 1 receptor 43 |
| chr3_+_77261334 | 3.21 |
ENSRNOT00000049527
|
LOC103690355
|
olfactory receptor 4C11-like |
| chr10_+_56824505 | 3.21 |
ENSRNOT00000067128
|
Slc16a11
|
solute carrier family 16, member 11 |
| chr7_+_117304742 | 3.16 |
ENSRNOT00000059599
|
Grina
|
glutamate ionotropic receptor NMDA type subunit associated protein 1 |
| chr2_+_57276919 | 3.15 |
ENSRNOT00000063899
|
RGD1310081
|
similar to hypothetical protein FLJ13231 |
| chr1_+_85112247 | 3.10 |
ENSRNOT00000087157
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr2_-_165739874 | 3.07 |
ENSRNOT00000014384
|
Kpna4
|
karyopherin subunit alpha 4 |
| chr3_+_75957592 | 3.04 |
ENSRNOT00000073455
|
Olr590
|
olfactory receptor 590 |
| chr3_-_102802338 | 3.03 |
ENSRNOT00000050334
|
Olr768
|
olfactory receptor 768 |
| chr8_+_117455262 | 3.01 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr1_-_149633029 | 2.99 |
ENSRNOT00000048861
|
LOC100910049
|
olfactory receptor 14A2-like |
| chr7_+_123531682 | 2.98 |
ENSRNOT00000010606
|
Wbp2nl
|
WBP2 N-terminal like |
| chr3_+_150761329 | 2.97 |
ENSRNOT00000067854
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr3_+_48626038 | 2.94 |
ENSRNOT00000009697
|
Gca
|
grancalcin |
| chr11_-_1437732 | 2.93 |
ENSRNOT00000037345
|
Csnka2ip
|
casein kinase 2, alpha prime interacting protein |
| chr11_-_54344615 | 2.92 |
ENSRNOT00000090307
|
Myh15
|
myosin, heavy chain 15 |
| chr7_+_140032132 | 2.91 |
ENSRNOT00000014316
|
Olr1107
|
olfactory receptor 1107 |
| chr2_-_195279218 | 2.89 |
ENSRNOT00000085565
|
RGD1559714
|
similar to TDPOZ3 |
| chr9_+_73528681 | 2.88 |
ENSRNOT00000084340
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr9_-_55673704 | 2.84 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr3_+_56125924 | 2.84 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr1_-_255557055 | 2.83 |
ENSRNOT00000033780
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr4_+_57925323 | 2.82 |
ENSRNOT00000085798
|
Cpa5
|
carboxypeptidase A5 |
| chr1_-_167588630 | 2.81 |
ENSRNOT00000050707
|
Olr39
|
olfactory receptor 39 |
| chr3_-_13865843 | 2.79 |
ENSRNOT00000025194
|
Rabepk
|
Rab9 effector protein with kelch motifs |
| chr7_-_58343137 | 2.75 |
ENSRNOT00000005310
ENSRNOT00000090560 |
Tmem19
|
transmembrane protein 19 |
| chr3_+_114918648 | 2.74 |
ENSRNOT00000089204
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr7_-_54855557 | 2.74 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chrX_+_123404518 | 2.74 |
ENSRNOT00000015085
|
Slc25a5
|
solute carrier family 25 member 5 |
| chr5_+_98387291 | 2.72 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr5_+_64476317 | 2.72 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr10_-_12312692 | 2.66 |
ENSRNOT00000073059
|
Olr1355
|
olfactory receptor 1355 |
| chr3_-_75089064 | 2.66 |
ENSRNOT00000046167
|
Olr542
|
olfactory receptor 542 |
| chr7_-_6691679 | 2.64 |
ENSRNOT00000075430
|
Olr959
|
olfactory receptor 959 |
| chr7_-_13404858 | 2.64 |
ENSRNOT00000060456
|
Olr1075
|
olfactory receptor 1075 |
| chrX_+_65796787 | 2.64 |
ENSRNOT00000076029
|
Gpr165
|
G protein-coupled receptor 165 |
| chr1_-_170404056 | 2.62 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr13_-_73704480 | 2.62 |
ENSRNOT00000005296
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr3_-_75235439 | 2.59 |
ENSRNOT00000046688
|
Olr552
|
olfactory receptor 552 |
| chr20_+_41266566 | 2.59 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr1_-_104166367 | 2.58 |
ENSRNOT00000092211
|
Csrp3
|
cysteine and glycine rich protein 3 |
| chr13_-_73704678 | 2.57 |
ENSRNOT00000005280
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr10_-_45940137 | 2.54 |
ENSRNOT00000004282
|
Olr1463
|
olfactory receptor 1463 |
| chr7_+_139762614 | 2.53 |
ENSRNOT00000031157
|
Ccdc184
|
coiled-coil domain containing 184 |
| chr5_-_62260089 | 2.53 |
ENSRNOT00000064858
ENSRNOT00000011714 |
Tbc1d2
|
TBC1 domain family, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nanog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 7.4 | 22.2 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 3.4 | 20.6 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 3.1 | 9.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 3.0 | 8.9 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 2.8 | 17.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 2.6 | 7.7 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) regulation of establishment of blood-brain barrier(GO:0090210) |
| 2.4 | 9.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.3 | 9.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.0 | 8.0 | GO:0098923 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 1.9 | 11.5 | GO:0008050 | female courtship behavior(GO:0008050) |
| 1.8 | 7.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.8 | 8.9 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 1.7 | 5.2 | GO:0019677 | NADP catabolic process(GO:0006742) NAD catabolic process(GO:0019677) |
| 1.7 | 6.9 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 1.6 | 6.5 | GO:1905231 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 1.6 | 9.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.5 | 6.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.5 | 6.0 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 1.4 | 9.8 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.4 | 4.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.3 | 3.8 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 1.1 | 13.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 3.3 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 1.0 | 4.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.0 | 7.9 | GO:0044598 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 1.0 | 3.8 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 1.0 | 9.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.9 | 2.8 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.9 | 4.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.9 | 11.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.9 | 2.6 | GO:0035995 | detection of muscle stretch(GO:0035995) regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.8 | 10.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.8 | 2.5 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.8 | 4.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.8 | 3.8 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.8 | 3.8 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.8 | 3.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.7 | 5.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.7 | 9.6 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.7 | 2.1 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.7 | 2.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.7 | 2.7 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.7 | 6.0 | GO:0051890 | regulation of cardioblast differentiation(GO:0051890) |
| 0.7 | 2.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.6 | 8.5 | GO:0097369 | sodium ion import(GO:0097369) |
| 0.6 | 4.8 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.6 | 3.6 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.6 | 2.4 | GO:0050904 | Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.6 | 1.8 | GO:0048749 | compound eye development(GO:0048749) |
| 0.6 | 8.2 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.6 | 22.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.6 | 1.7 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.6 | 7.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.5 | 5.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.5 | 2.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.5 | 2.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 2.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.5 | 3.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.5 | 14.3 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.5 | 3.6 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.5 | 3.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.4 | 1.3 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) positive regulation of cytolysis(GO:0045919) cellular response to iron(II) ion(GO:0071282) |
| 0.4 | 1.3 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.4 | 1.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 4.4 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.4 | 1.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.4 | 0.7 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.3 | 1.4 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.3 | 11.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 2.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 2.7 | GO:0015866 | ADP transport(GO:0015866) |
| 0.3 | 4.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 3.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 1.7 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.3 | 2.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.3 | 2.8 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.3 | 8.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.3 | 9.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.3 | 6.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 2.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 3.9 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 5.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 1.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 2.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 21.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.2 | 3.8 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.2 | 1.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 0.9 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.2 | 4.9 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 0.7 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 | 227.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 4.4 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 3.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 0.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 3.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 7.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 7.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 1.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.2 | 4.0 | GO:0031572 | mitotic G2 DNA damage checkpoint(GO:0007095) G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 1.4 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.2 | 1.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 0.9 | GO:2000323 | regulation of type B pancreatic cell development(GO:2000074) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 3.8 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.1 | 4.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 2.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 3.0 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 5.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 3.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 2.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 9.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 2.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 5.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 2.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 2.0 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 2.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.9 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.3 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 | 0.8 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 13.7 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.1 | 2.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 4.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 10.4 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 0.1 | GO:0014901 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.1 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 3.4 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 2.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.4 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.9 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 1.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 4.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 2.2 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 2.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.8 | 11.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.5 | 6.1 | GO:0071914 | prominosome(GO:0071914) |
| 1.3 | 13.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 1.3 | 7.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.0 | 6.0 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.0 | 3.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.0 | 17.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.8 | 4.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.7 | 3.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.7 | 2.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.6 | 10.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.6 | 6.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.6 | 8.8 | GO:0060091 | kinocilium(GO:0060091) |
| 0.5 | 7.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.5 | 6.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.5 | 2.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.5 | 5.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 8.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.4 | 3.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 7.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 32.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 14.1 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 2.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 4.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 4.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 0.7 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 10.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 10.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 1.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 2.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 17.6 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 4.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 0.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 7.9 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 2.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 3.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 7.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 3.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 4.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 2.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 12.1 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 4.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 32.2 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 8.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 6.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 8.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 3.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 4.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.9 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 3.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 16.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 4.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 6.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 4.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 124.9 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 4.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 8.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.8 | GO:0044429 | mitochondrial part(GO:0044429) |
| 0.0 | 0.3 | GO:0030016 | myofibril(GO:0030016) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 6.4 | 32.0 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 3.1 | 9.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 2.6 | 7.9 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 2.6 | 7.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 2.4 | 9.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 2.0 | 14.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.6 | 8.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.6 | 7.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.5 | 6.1 | GO:0016160 | amylase activity(GO:0016160) |
| 1.4 | 17.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 1.3 | 6.5 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 1.3 | 5.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 1.3 | 3.8 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.3 | 3.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 1.3 | 3.8 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 1.3 | 3.8 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 1.2 | 10.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.0 | 11.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.0 | 8.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 1.0 | 3.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.9 | 8.9 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.9 | 7.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.9 | 13.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.8 | 11.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.8 | 10.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.8 | 5.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.8 | 5.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.7 | 5.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.7 | 4.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.7 | 2.7 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.6 | 2.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 3.8 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.6 | 6.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.6 | 17.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 3.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 3.7 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.5 | 4.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.5 | 6.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 14.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 2.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.5 | 21.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.5 | 22.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.5 | 1.4 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.5 | 3.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.4 | 1.7 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.4 | 12.8 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.4 | 34.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.4 | 10.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 2.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 6.0 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.4 | 10.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.4 | 1.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.3 | 6.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.3 | 13.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 3.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.3 | 5.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 5.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.3 | 1.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.3 | 2.1 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.3 | 10.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 1.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 4.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 5.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 2.0 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 2.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 2.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 227.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 6.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.5 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 3.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 4.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 8.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 14.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 2.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 8.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 3.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.7 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 3.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 3.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 5.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 4.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 1.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 2.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 2.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 2.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 6.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 2.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.5 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.6 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 12.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 11.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 13.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 7.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 6.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 7.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 4.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 20.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 1.8 | 5.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 1.0 | 6.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 1.0 | 9.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.8 | 89.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.6 | 17.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.6 | 7.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 2.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.4 | 10.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 11.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 4.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 4.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 6.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 4.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 7.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 3.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 3.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 3.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 4.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 11.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 5.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 5.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 3.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 5.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 2.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 6.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 5.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.3 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 2.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 7.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |