Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
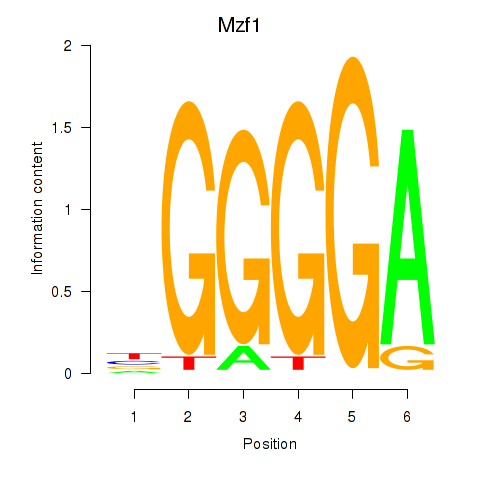
Results for Mzf1
Z-value: 0.63
Transcription factors associated with Mzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mzf1
|
ENSRNOG00000027550 | myeloid zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mzf1 | rn6_v1_chr1_+_65522118_65522118 | -0.13 | 1.7e-02 | Click! |
Activity profile of Mzf1 motif
Sorted Z-values of Mzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_70072156 | 17.51 |
ENSRNOT00000046667
|
Calml3
|
calmodulin-like 3 |
| chr5_-_24631679 | 12.79 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr2_+_206064179 | 11.57 |
ENSRNOT00000025953
|
Syt6
|
synaptotagmin 6 |
| chr7_+_70364813 | 11.41 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr2_+_206064394 | 11.17 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr1_+_144831523 | 10.77 |
ENSRNOT00000039748
|
Mex3b
|
mex-3 RNA binding family member B |
| chr5_+_154522119 | 10.27 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr17_+_43627930 | 10.16 |
ENSRNOT00000081719
|
LOC102549061
|
histone H2B type 1-N-like |
| chr8_+_47674321 | 9.97 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr19_+_54060622 | 8.83 |
ENSRNOT00000023607
|
Gse1
|
Gse1 coiled-coil protein |
| chr3_+_16753703 | 8.51 |
ENSRNOT00000077741
|
AABR07051548.2
|
|
| chr7_+_142912316 | 8.41 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_82480195 | 8.08 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr7_-_70969905 | 8.04 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr12_-_9331195 | 8.01 |
ENSRNOT00000044134
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr2_+_188495629 | 8.01 |
ENSRNOT00000027828
|
Fam189b
|
family with sequence similarity 189, member B |
| chr3_-_148722710 | 7.80 |
ENSRNOT00000090919
ENSRNOT00000068592 |
Plagl2
|
PLAG1 like zinc finger 2 |
| chr2_-_207300854 | 7.29 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr20_+_5527181 | 7.07 |
ENSRNOT00000091364
|
Phf1
|
PHD finger protein 1 |
| chr3_+_124068865 | 6.61 |
ENSRNOT00000079264
|
Smox
|
spermine oxidase |
| chr20_+_46429222 | 6.38 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr10_+_97212432 | 6.22 |
ENSRNOT00000088599
|
Axin2
|
axin 2 |
| chr20_+_6923489 | 6.19 |
ENSRNOT00000093373
ENSRNOT00000000632 |
Pi16
|
peptidase inhibitor 16 |
| chr3_-_7795758 | 6.10 |
ENSRNOT00000045919
|
Ntng2
|
netrin G2 |
| chr3_-_150064438 | 5.94 |
ENSRNOT00000086933
|
E2f1
|
E2F transcription factor 1 |
| chr5_-_151977636 | 5.73 |
ENSRNOT00000009173
|
Arid1a
|
AT-rich interaction domain 1A |
| chr2_-_219458271 | 5.65 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr20_+_5374985 | 5.63 |
ENSRNOT00000052270
|
RT1-A2
|
RT1 class Ia, locus A2 |
| chr10_+_94280703 | 5.59 |
ENSRNOT00000088656
|
Map3k3
|
mitogen activated protein kinase kinase kinase 3 |
| chr6_-_21600451 | 5.36 |
ENSRNOT00000047674
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_196334626 | 5.27 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr7_+_119482272 | 5.18 |
ENSRNOT00000009544
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr10_-_56154548 | 5.04 |
ENSRNOT00000090809
|
Dnah2
|
dynein, axonemal, heavy chain 2 |
| chr4_-_115157263 | 4.77 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr15_+_33555640 | 4.77 |
ENSRNOT00000021096
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr11_-_65209268 | 4.76 |
ENSRNOT00000077612
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr19_+_26142720 | 4.72 |
ENSRNOT00000005270
|
RGD1564093
|
similar to RIKEN cDNA 2310036O22 |
| chr20_-_1818800 | 4.70 |
ENSRNOT00000000990
|
RT1-M3-1
|
RT1 class Ib, locus M3, gene 1 |
| chr1_+_80000165 | 4.69 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr4_+_102665529 | 4.61 |
ENSRNOT00000082333
|
AABR07061005.1
|
|
| chr14_+_108007724 | 4.46 |
ENSRNOT00000014062
|
Xpo1
|
exportin 1 |
| chr20_-_5140304 | 4.44 |
ENSRNOT00000092646
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr12_+_46615649 | 4.41 |
ENSRNOT00000086531
ENSRNOT00000089494 |
Bicdl1
|
BICD family like cargo adaptor 1 |
| chr4_-_82300503 | 4.36 |
ENSRNOT00000071568
|
Hoxa11
|
homeobox A11 |
| chr9_+_81518584 | 4.30 |
ENSRNOT00000084309
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr10_-_64862268 | 4.28 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chr1_+_198214797 | 4.03 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr10_+_56591292 | 4.02 |
ENSRNOT00000023379
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr17_-_54808483 | 3.98 |
ENSRNOT00000075683
|
AABR07028049.1
|
|
| chr12_+_21721837 | 3.97 |
ENSRNOT00000073353
|
Mepce
|
methylphosphate capping enzyme |
| chr15_-_45927804 | 3.93 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr4_+_82214342 | 3.57 |
ENSRNOT00000066360
|
Hoxa11-as
|
homeobox A11, opposite strand |
| chr3_-_156340913 | 3.53 |
ENSRNOT00000021452
|
Mafb
|
MAF bZIP transcription factor B |
| chr12_-_21760292 | 3.49 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr2_-_196415530 | 3.45 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr9_+_82033543 | 3.44 |
ENSRNOT00000023439
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr4_+_8256611 | 3.43 |
ENSRNOT00000061894
|
AABR07059198.1
|
|
| chr6_+_109300433 | 3.42 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr1_+_72636959 | 3.36 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr7_-_144880092 | 3.35 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr3_-_119135391 | 3.34 |
ENSRNOT00000045443
|
Gabpb1
|
GA binding protein transcription factor, beta subunit 1 |
| chr2_+_113984646 | 3.23 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr10_-_85124644 | 3.21 |
ENSRNOT00000012376
|
Kpnb1
|
karyopherin subunit beta 1 |
| chr4_-_152835182 | 3.20 |
ENSRNOT00000036721
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr13_-_41738622 | 3.15 |
ENSRNOT00000004520
ENSRNOT00000084552 |
Actr3
|
ARP3 actin related protein 3 homolog |
| chr20_+_4993560 | 3.14 |
ENSRNOT00000081628
ENSRNOT00000087861 ENSRNOT00000001160 |
Vars
|
valyl-tRNA synthetase |
| chr6_+_132702448 | 3.12 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr14_-_35652709 | 3.04 |
ENSRNOT00000003080
|
Gsx2
|
GS homeobox 2 |
| chr2_+_210381829 | 3.03 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr2_+_205160405 | 2.97 |
ENSRNOT00000035605
|
Tspan2
|
tetraspanin 2 |
| chr1_+_183892841 | 2.91 |
ENSRNOT00000015498
|
Pde3b
|
phosphodiesterase 3B |
| chr10_-_56591364 | 2.81 |
ENSRNOT00000032481
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr3_-_46051096 | 2.80 |
ENSRNOT00000081302
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr13_+_51126459 | 2.74 |
ENSRNOT00000042046
ENSRNOT00000087240 |
Myog
|
myogenin |
| chr10_+_68588789 | 2.69 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr7_-_3074359 | 2.53 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr8_-_111850075 | 2.51 |
ENSRNOT00000082097
|
Cdv3
|
CDV3 homolog |
| chr10_-_110182291 | 2.48 |
ENSRNOT00000015178
ENSRNOT00000054936 |
Csnk1d
|
casein kinase 1, delta |
| chr5_+_144160108 | 2.45 |
ENSRNOT00000064972
|
Eva1b
|
eva-1 homolog B |
| chr10_+_56627411 | 2.39 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_-_52894832 | 2.34 |
ENSRNOT00000016471
|
T
|
T brachyury transcription factor |
| chr4_+_145017608 | 2.33 |
ENSRNOT00000066723
|
Setd5
|
SET domain containing 5 |
| chr3_+_170550314 | 2.32 |
ENSRNOT00000006991
|
Tfap2c
|
transcription factor AP-2 gamma |
| chr2_-_208152179 | 2.31 |
ENSRNOT00000068152
|
Ddx20
|
DEAD-box helicase 20 |
| chrX_-_1856258 | 2.25 |
ENSRNOT00000079980
|
RGD1564855
|
similar to RuvB-like protein 1 |
| chr1_-_154216340 | 2.23 |
ENSRNOT00000024082
|
Eed
|
embryonic ectoderm development |
| chr13_-_45040593 | 2.17 |
ENSRNOT00000004908
|
Lct
|
lactase |
| chr1_-_101388125 | 2.14 |
ENSRNOT00000028176
ENSRNOT00000049957 |
Snrnp70
|
small nuclear ribonucleoprotein U1 subunit 70 |
| chr18_+_65814026 | 2.13 |
ENSRNOT00000016112
|
Mbd2
|
methyl-CpG binding domain protein 2 |
| chr10_+_86860685 | 2.10 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr5_+_147476221 | 2.04 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr12_-_21728678 | 2.01 |
ENSRNOT00000035372
|
LOC100910581
|
protein phosphatase 1 regulatory subunit 35-like |
| chr9_+_16612433 | 2.00 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr10_+_86157608 | 1.98 |
ENSRNOT00000082668
ENSRNOT00000008256 |
Cdk12
|
cyclin-dependent kinase 12 |
| chr17_+_36334589 | 1.97 |
ENSRNOT00000081368
|
E2f3
|
E2F transcription factor 3 |
| chr3_+_112228919 | 1.95 |
ENSRNOT00000011761
|
Capn3
|
calpain 3 |
| chr6_+_10533151 | 1.94 |
ENSRNOT00000020822
|
Rhoq
|
ras homolog family member Q |
| chr12_+_40510378 | 1.93 |
ENSRNOT00000001817
|
Mapkapk5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr2_-_46544457 | 1.90 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr6_+_128973956 | 1.90 |
ENSRNOT00000075399
|
Fam181a
|
family with sequence similarity 181, member A |
| chr7_-_140546908 | 1.85 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr18_-_6474990 | 1.81 |
ENSRNOT00000061504
|
Kctd1
|
potassium channel tetramerization domain containing 1 |
| chr15_+_23665202 | 1.78 |
ENSRNOT00000089226
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr10_-_64657089 | 1.77 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr5_-_50221095 | 1.76 |
ENSRNOT00000012164
|
RGD1563056
|
similar to hypothetical protein |
| chr14_-_86868598 | 1.76 |
ENSRNOT00000087212
|
Nacad
|
NAC alpha domain containing |
| chr3_-_8659102 | 1.74 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr1_+_278198179 | 1.71 |
ENSRNOT00000023247
|
Fam160b1
|
family with sequence similarity 160, member B1 |
| chr3_+_112228720 | 1.70 |
ENSRNOT00000079079
|
Capn3
|
calpain 3 |
| chr4_-_118472179 | 1.69 |
ENSRNOT00000023856
|
Mxd1
|
max dimerization protein 1 |
| chr15_-_52210746 | 1.68 |
ENSRNOT00000046054
|
Bmp1
|
bone morphogenetic protein 1 |
| chr1_+_197999037 | 1.65 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr6_+_108831108 | 1.65 |
ENSRNOT00000038594
|
LOC103692678
|
U6 snRNA-associated Sm-like protein LSm5 pseudogene |
| chr1_+_44446765 | 1.61 |
ENSRNOT00000030132
|
Cldn20
|
claudin 20 |
| chr15_+_12827707 | 1.61 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr4_-_57625147 | 1.60 |
ENSRNOT00000074649
ENSRNOT00000078961 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr10_-_56365084 | 1.53 |
ENSRNOT00000068013
|
Polr2a
|
RNA polymerase II subunit A |
| chr4_+_98648545 | 1.52 |
ENSRNOT00000008451
|
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr1_-_72377434 | 1.51 |
ENSRNOT00000022193
|
Zfp524
|
zinc finger protein 524 |
| chr5_+_140979435 | 1.51 |
ENSRNOT00000056592
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr1_+_199225100 | 1.50 |
ENSRNOT00000088606
|
Setd1a
|
SET domain containing 1A |
| chr1_+_197999336 | 1.46 |
ENSRNOT00000023555
|
Apobr
|
apolipoprotein B receptor |
| chr5_-_146069670 | 1.44 |
ENSRNOT00000072793
|
LOC682102
|
hypothetical protein LOC682102 |
| chr4_-_56114254 | 1.40 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr8_-_63291966 | 1.40 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr5_-_144556889 | 1.37 |
ENSRNOT00000035373
|
Ago4
|
argonaute 4, RISC catalytic component |
| chr12_+_22229079 | 1.36 |
ENSRNOT00000001911
|
Gnb2
|
G protein subunit beta 2 |
| chr15_+_26033791 | 1.30 |
ENSRNOT00000020074
|
Naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr8_+_48805684 | 1.29 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr7_+_142869752 | 1.25 |
ENSRNOT00000009979
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chr20_-_1984737 | 1.23 |
ENSRNOT00000040232
ENSRNOT00000051634 ENSRNOT00000079445 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr3_-_57119001 | 1.19 |
ENSRNOT00000083171
|
Tlk1
|
tousled-like kinase 1 |
| chr2_+_188844073 | 1.18 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr10_-_56444847 | 1.12 |
ENSRNOT00000056872
ENSRNOT00000092662 |
Nlgn2
|
neuroligin 2 |
| chr4_+_159622404 | 1.11 |
ENSRNOT00000078299
|
Fgf23
|
fibroblast growth factor 23 |
| chr1_+_280633938 | 1.09 |
ENSRNOT00000012564
|
Emx2
|
empty spiracles homeobox 2 |
| chr5_-_59085676 | 1.08 |
ENSRNOT00000068105
|
Msmp
|
microseminoprotein, prostate associated |
| chr15_-_12513931 | 1.08 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chr18_+_30890869 | 1.08 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr1_+_87180624 | 0.92 |
ENSRNOT00000056933
ENSRNOT00000027987 |
LOC103689986
|
protein YIF1B |
| chr10_-_85684138 | 0.90 |
ENSRNOT00000017989
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase type 2 beta |
| chr16_-_36373546 | 0.89 |
ENSRNOT00000079552
|
Hand2
|
heart and neural crest derivatives expressed transcript 2 |
| chr9_+_82596355 | 0.88 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr2_-_77632628 | 0.86 |
ENSRNOT00000073915
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr2_+_198762138 | 0.84 |
ENSRNOT00000028811
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chrX_-_157095274 | 0.84 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr4_+_66290389 | 0.82 |
ENSRNOT00000008557
|
Luc7l2
|
LUC7-like 2 pre-mRNA splicing factor |
| chr10_-_86144880 | 0.80 |
ENSRNOT00000067901
|
Med1
|
mediator complex subunit 1 |
| chr20_-_1980101 | 0.76 |
ENSRNOT00000084582
ENSRNOT00000085050 ENSRNOT00000082545 ENSRNOT00000088396 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr9_-_10991820 | 0.76 |
ENSRNOT00000073011
ENSRNOT00000081441 |
Hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr5_-_58113553 | 0.74 |
ENSRNOT00000046589
|
Arid3c
|
AT-rich interaction domain 3C |
| chrX_-_63204530 | 0.73 |
ENSRNOT00000076175
|
Zfx
|
zinc finger protein X-linked |
| chr9_+_20264418 | 0.71 |
ENSRNOT00000051841
|
Atn1
|
atrophin 1 |
| chr7_-_36499784 | 0.70 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr2_-_199354793 | 0.66 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr10_+_84200880 | 0.64 |
ENSRNOT00000011213
|
Hoxb2
|
homeobox B2 |
| chr7_-_140454268 | 0.62 |
ENSRNOT00000081468
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr10_-_65963932 | 0.62 |
ENSRNOT00000011726
|
Nlk
|
nemo like kinase |
| chr6_+_98284170 | 0.60 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr7_+_58366192 | 0.60 |
ENSRNOT00000081891
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr20_-_6961162 | 0.57 |
ENSRNOT00000000635
|
Mtch1
|
mitochondrial carrier 1 |
| chr2_+_113984824 | 0.55 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr10_-_85574889 | 0.55 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr20_+_5050327 | 0.49 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr17_-_27665266 | 0.49 |
ENSRNOT00000060218
|
Rreb1
|
ras responsive element binding protein 1 |
| chr5_+_59080765 | 0.47 |
ENSRNOT00000021888
ENSRNOT00000064419 |
Rgp1
|
RGP1 homolog, RAB6A GEF complex partner 1 |
| chr1_-_199865548 | 0.45 |
ENSRNOT00000027463
|
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr2_-_198719202 | 0.36 |
ENSRNOT00000028801
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr4_-_155116154 | 0.28 |
ENSRNOT00000020529
|
Phc1
|
polyhomeotic homolog 1 |
| chr2_+_188748359 | 0.25 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr17_-_90217786 | 0.25 |
ENSRNOT00000073534
|
Impad1
|
inositol monophosphatase domain containing 1 |
| chr7_+_25919867 | 0.24 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr20_-_1099336 | 0.21 |
ENSRNOT00000071766
|
LOC100362856
|
olfactory receptor-like protein-like |
| chr11_-_83926524 | 0.16 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_+_7279340 | 0.16 |
ENSRNOT00000017029
|
Ak8
|
adenylate kinase 8 |
| chr5_+_169338097 | 0.13 |
ENSRNOT00000014035
|
Hes2
|
hes family bHLH transcription factor 2 |
| chr9_+_17841410 | 0.12 |
ENSRNOT00000031706
|
Tmem151b
|
transmembrane protein 151B |
| chr1_+_199360645 | 0.12 |
ENSRNOT00000026527
|
Kat8
|
lysine acetyltransferase 8 |
| chr14_+_4125380 | 0.12 |
ENSRNOT00000002882
|
Zfp644
|
zinc finger protein 644 |
| chr10_+_86711240 | 0.08 |
ENSRNOT00000012812
|
Msl1
|
male specific lethal 1 homolog |
| chr6_+_127207732 | 0.01 |
ENSRNOT00000071976
|
RGD1309028
|
similar to RIKEN cDNA A830059I20 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 22.7 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 2.7 | 16.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 2.7 | 8.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 2.7 | 8.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 2.2 | 10.8 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 2.1 | 6.2 | GO:2000054 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) regulation of centromeric sister chromatid cohesion(GO:0070602) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.9 | 5.7 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 1.8 | 8.8 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 1.7 | 8.7 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.6 | 4.7 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 1.4 | 8.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.2 | 3.6 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 1.2 | 4.8 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 1.2 | 9.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 1.1 | 6.6 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 1.1 | 4.3 | GO:2000814 | positive regulation of barbed-end actin filament capping(GO:2000814) |
| 1.1 | 3.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 1.1 | 6.4 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 1.1 | 12.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 1.1 | 3.2 | GO:1904170 | meiotic chromosome movement towards spindle pole(GO:0016344) regulation of bleb assembly(GO:1904170) |
| 1.0 | 3.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 1.0 | 8.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.9 | 3.5 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.8 | 3.1 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.8 | 6.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.7 | 5.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.7 | 3.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.7 | 2.7 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.6 | 0.6 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 4 development(GO:0021570) |
| 0.6 | 4.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.6 | 1.8 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.6 | 2.3 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) notochord formation(GO:0014028) |
| 0.6 | 4.0 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.5 | 1.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 1.5 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.5 | 2.4 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.4 | 3.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.4 | 4.7 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.4 | 3.4 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.4 | 3.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.4 | 1.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.4 | 3.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.4 | 1.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) response to sodium phosphate(GO:1904383) |
| 0.4 | 1.9 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.4 | 11.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.4 | 2.2 | GO:0010045 | response to nickel cation(GO:0010045) |
| 0.3 | 3.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 4.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 7.8 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.3 | 2.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.3 | 2.3 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.3 | 2.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 2.5 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.3 | 2.1 | GO:0060591 | positive regulation of T-helper 2 cell differentiation(GO:0045630) chondroblast differentiation(GO:0060591) |
| 0.3 | 4.4 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.3 | 0.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 5.4 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.2 | 4.0 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 | 0.9 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.2 | 0.7 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.2 | 1.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 2.5 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.2 | 1.9 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.2 | 3.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 2.1 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.2 | 1.5 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.1 | 0.8 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 2.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 2.9 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 1.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 1.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.8 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 1.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 10.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 2.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.4 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 | 1.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.7 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 1.9 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 3.0 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 2.2 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 2.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 5.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 1.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 3.1 | GO:0006641 | triglyceride metabolic process(GO:0006641) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.7 | GO:0048599 | oocyte development(GO:0048599) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 1.0 | 5.9 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.0 | 19.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.7 | 4.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.7 | 4.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.7 | 3.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.7 | 2.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.7 | 2.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.6 | 4.7 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.6 | 5.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 2.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 4.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.5 | 5.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.5 | 5.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.5 | 4.8 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.5 | 9.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 5.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 3.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 2.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 1.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.3 | 4.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 2.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 2.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 3.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 3.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 3.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 19.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.9 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 5.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 3.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 10.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 9.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 8.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 7.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 4.0 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) |
| 0.1 | 0.8 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 18.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 3.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 16.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 5.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 11.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 5.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 1.6 | 8.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.2 | 4.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.0 | 3.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.7 | 2.2 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 0.7 | 4.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.6 | 1.9 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.6 | 3.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.6 | 4.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.6 | 4.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 10.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.5 | 2.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 10.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.5 | 8.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 4.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 6.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.4 | 5.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 2.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.4 | 11.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 6.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 8.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 1.9 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 3.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 2.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 0.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 16.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 2.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.3 | 3.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 1.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 4.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 10.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 2.1 | GO:1990446 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.2 | 9.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 5.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 1.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 3.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 1.5 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 2.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.2 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.2 | 1.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 11.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 30.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 7.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 3.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 4.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 5.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 4.0 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.1 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.2 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 12.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 5.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 4.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 3.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 6.0 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 2.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.8 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 13.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.4 | 8.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 18.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.3 | 4.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 3.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 11.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 5.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 8.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 6.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 7.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 4.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 18.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 8.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 14.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.4 | 6.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 2.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 5.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 7.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 3.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 4.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 3.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 4.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 5.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 5.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 2.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 3.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |