Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
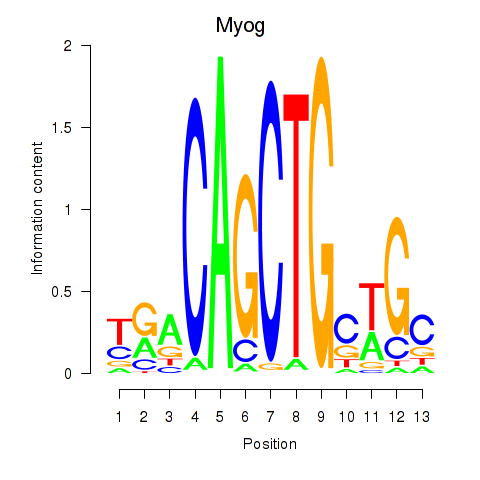
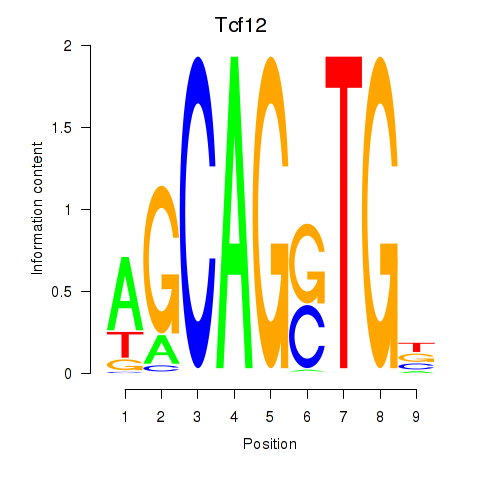
Results for Myog_Tcf12
Z-value: 2.20
Transcription factors associated with Myog_Tcf12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myog
|
ENSRNOG00000030743 | myogenin |
|
Tcf12
|
ENSRNOG00000057754 | transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myog | rn6_v1_chr13_+_51126459_51126459 | 0.27 | 1.5e-06 | Click! |
| Tcf12 | rn6_v1_chr8_-_78655856_78655856 | -0.12 | 2.8e-02 | Click! |
Activity profile of Myog_Tcf12 motif
Sorted Z-values of Myog_Tcf12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_168018410 | 165.44 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr19_-_11326139 | 112.40 |
ENSRNOT00000025669
|
Mt3
|
metallothionein 3 |
| chr10_-_8654892 | 104.78 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr2_-_231521052 | 102.96 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr9_+_14529218 | 88.86 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr1_-_81881549 | 72.91 |
ENSRNOT00000027497
|
Atp1a3
|
ATPase Na+/K+ transporting subunit alpha 3 |
| chr10_-_15928169 | 69.43 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr10_+_92628356 | 69.15 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr10_-_87578854 | 65.81 |
ENSRNOT00000065619
|
LOC680160
|
similar to keratin associated protein 4-7 |
| chr17_+_81922329 | 65.15 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr18_+_52215682 | 61.30 |
ENSRNOT00000037901
|
Megf10
|
multiple EGF-like domains 10 |
| chr18_+_56193978 | 61.18 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr10_-_8498422 | 60.98 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr3_+_11114551 | 59.73 |
ENSRNOT00000013507
|
Plpp7
|
phospholipid phosphatase 7 |
| chr14_-_86297623 | 59.58 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chrX_+_105239840 | 59.47 |
ENSRNOT00000039864
|
Drp2
|
dystrophin related protein 2 |
| chr2_-_21698937 | 58.29 |
ENSRNOT00000080165
|
AABR07007642.1
|
|
| chr11_+_72705129 | 56.43 |
ENSRNOT00000073330
|
Apod
|
apolipoprotein D |
| chr1_-_73753128 | 56.23 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr4_-_147163467 | 56.20 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr1_-_102106127 | 55.86 |
ENSRNOT00000028685
|
Kcnj11
|
potassium voltage-gated channel subfamily J member 11 |
| chrX_+_23081125 | 53.49 |
ENSRNOT00000071639
|
AABR07037520.1
|
|
| chr9_-_82699551 | 51.99 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr16_+_81616604 | 51.54 |
ENSRNOT00000026392
ENSRNOT00000057740 |
Adprhl1
Grtp1
|
ADP-ribosylhydrolase like 1 growth hormone regulated TBC protein 1 |
| chr2_+_11658568 | 49.13 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_80331626 | 48.65 |
ENSRNOT00000022577
|
AABR07002677.1
|
|
| chr2_+_210977938 | 48.26 |
ENSRNOT00000074725
|
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr7_-_50278842 | 47.59 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr1_+_222310920 | 47.54 |
ENSRNOT00000091465
|
Macrod1
|
MACRO domain containing 1 |
| chr1_+_222311253 | 47.26 |
ENSRNOT00000028749
|
Macrod1
|
MACRO domain containing 1 |
| chr5_-_109651730 | 47.00 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr1_+_79989019 | 46.53 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr14_+_60764409 | 46.35 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr17_+_1679627 | 45.85 |
ENSRNOT00000025801
|
Habp4
|
hyaluronan binding protein 4 |
| chr10_+_70262361 | 45.79 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr3_+_41019898 | 45.29 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr5_+_144634143 | 44.71 |
ENSRNOT00000075558
|
RGD1563072
|
similar to hypothetical protein FLJ38984 |
| chr10_-_95934345 | 44.33 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr4_+_51553454 | 43.14 |
ENSRNOT00000073466
|
LOC100909784
|
leiomodin-2-like |
| chr10_-_94557764 | 42.16 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chr2_+_242882306 | 41.68 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr1_-_89560719 | 41.65 |
ENSRNOT00000028653
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr13_+_52662996 | 40.92 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr15_+_110114148 | 40.89 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr3_-_161246351 | 40.67 |
ENSRNOT00000020348
|
Tnnc2
|
troponin C2, fast skeletal type |
| chr1_+_78800754 | 40.55 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr4_+_51661654 | 40.52 |
ENSRNOT00000087359
ENSRNOT00000072470 |
LOC100909784
|
leiomodin-2-like |
| chr1_+_80321585 | 39.90 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr4_-_157381105 | 39.82 |
ENSRNOT00000021670
|
Gpr162
|
G protein-coupled receptor 162 |
| chr12_+_24473981 | 39.53 |
ENSRNOT00000001973
|
Fzd9
|
frizzled class receptor 9 |
| chr6_-_147172813 | 39.38 |
ENSRNOT00000066545
|
Itgb8
|
integrin subunit beta 8 |
| chr9_+_82700468 | 39.03 |
ENSRNOT00000027227
|
Inha
|
inhibin alpha subunit |
| chr9_-_55673704 | 38.63 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr13_+_85818427 | 38.48 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr19_-_43596801 | 38.32 |
ENSRNOT00000025625
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr9_-_93735636 | 38.28 |
ENSRNOT00000025582
|
Nppc
|
natriuretic peptide C |
| chr17_+_13670520 | 38.25 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr1_-_89560469 | 38.19 |
ENSRNOT00000079091
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr5_+_1417478 | 38.07 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chrX_+_86126157 | 37.90 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr3_+_35014538 | 37.77 |
ENSRNOT00000006341
|
Kif5c
|
kinesin family member 5C |
| chr7_-_115910522 | 37.23 |
ENSRNOT00000076998
ENSRNOT00000067442 |
Arc
|
activity-regulated cytoskeleton-associated protein |
| chr5_-_160179978 | 37.12 |
ENSRNOT00000022820
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr10_-_62699723 | 37.06 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr1_-_215846911 | 36.67 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr1_+_142883040 | 36.58 |
ENSRNOT00000015898
|
Alpk3
|
alpha-kinase 3 |
| chr6_+_76349362 | 36.52 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr9_+_82556573 | 36.44 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr8_-_23063041 | 36.36 |
ENSRNOT00000018416
|
Elavl3
|
ELAV like RNA binding protein 3 |
| chr1_+_215609645 | 36.31 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr1_-_78851719 | 35.89 |
ENSRNOT00000022603
|
Calm2
|
calmodulin 2 |
| chr2_-_257484587 | 35.84 |
ENSRNOT00000083344
|
Nexn
|
nexilin (F actin binding protein) |
| chr7_-_2786856 | 35.77 |
ENSRNOT00000047530
ENSRNOT00000086939 |
Coq10a
|
coenzyme Q10A |
| chr18_-_37245809 | 35.67 |
ENSRNOT00000079585
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_31055453 | 34.94 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr20_-_4823475 | 34.92 |
ENSRNOT00000082536
ENSRNOT00000001114 |
Atp6v1g2
|
ATPase H+ transporting V1 subunit G2 |
| chr8_+_63600663 | 34.81 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr9_+_82120059 | 34.77 |
ENSRNOT00000057368
|
Cdk5r2
|
cyclin-dependent kinase 5 regulatory subunit 2 |
| chr4_+_113968995 | 34.59 |
ENSRNOT00000079511
|
Rtkn
|
rhotekin |
| chr8_-_84632817 | 34.54 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr17_-_80320681 | 34.39 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr8_-_21831668 | 34.34 |
ENSRNOT00000027897
|
Col5a3
|
collagen type V alpha 3 chain |
| chr3_-_102151489 | 34.33 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr4_-_129619142 | 34.25 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr13_-_42263024 | 34.03 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr1_+_220416018 | 33.68 |
ENSRNOT00000027233
|
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chr3_-_105279462 | 33.63 |
ENSRNOT00000010679
|
Scg5
|
secretogranin V |
| chr5_-_59025631 | 33.38 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chrX_+_110404022 | 33.28 |
ENSRNOT00000041044
|
Mum1l1
|
MUM1 like 1 |
| chr12_-_6740714 | 33.26 |
ENSRNOT00000001205
|
Medag
|
mesenteric estrogen-dependent adipogenesis |
| chr15_+_23792931 | 33.02 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chrX_-_136807885 | 32.74 |
ENSRNOT00000010325
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr15_+_19547871 | 32.35 |
ENSRNOT00000036235
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr2_-_188138177 | 32.03 |
ENSRNOT00000027474
|
Syt11
|
synaptotagmin 11 |
| chr20_+_5008508 | 31.82 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr3_-_2689084 | 30.97 |
ENSRNOT00000020926
|
Ptgds
|
prostaglandin D2 synthase |
| chr5_+_154112076 | 30.79 |
ENSRNOT00000044969
|
Myom3
|
myomesin 3 |
| chr9_+_9961021 | 30.68 |
ENSRNOT00000075767
|
Tubb4a
|
tubulin, beta 4A class IVa |
| chr1_-_265573117 | 30.64 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr13_-_82005741 | 30.41 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr2_-_172361779 | 30.31 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr18_+_16650806 | 30.10 |
ENSRNOT00000093679
ENSRNOT00000041961 ENSRNOT00000093140 |
Fhod3
|
formin homology 2 domain containing 3 |
| chr20_+_3556975 | 29.91 |
ENSRNOT00000089417
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr4_+_7076759 | 29.68 |
ENSRNOT00000066598
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr11_+_27208564 | 29.50 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr7_-_12274803 | 29.44 |
ENSRNOT00000049146
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr3_-_147143576 | 29.16 |
ENSRNOT00000091811
ENSRNOT00000012727 |
Snph
|
syntaphilin |
| chr13_-_82006005 | 29.10 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr14_-_112946875 | 28.76 |
ENSRNOT00000081981
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr4_+_9966891 | 28.65 |
ENSRNOT00000086609
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr4_-_117268178 | 28.53 |
ENSRNOT00000043201
ENSRNOT00000084049 |
Fbxo41
|
F-box protein 41 |
| chr12_-_36398206 | 28.48 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr2_-_115836846 | 28.26 |
ENSRNOT00000014359
|
Cldn11
|
claudin 11 |
| chr1_+_167197549 | 27.95 |
ENSRNOT00000027427
|
Art1
|
ADP-ribosyltransferase 1 |
| chr1_-_31122093 | 27.65 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chr1_+_101884276 | 27.62 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr15_-_90175802 | 27.54 |
ENSRNOT00000013342
|
Spry2
|
sprouty RTK signaling antagonist 2 |
| chr4_+_84597323 | 27.29 |
ENSRNOT00000074054
ENSRNOT00000012755 |
Wipf3
|
WAS/WASL interacting protein family, member 3 |
| chr13_-_39643361 | 26.89 |
ENSRNOT00000003527
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr16_+_34795971 | 26.84 |
ENSRNOT00000043510
|
LOC100912321
|
myeloid-associated differentiation marker-like |
| chr8_+_27777179 | 26.75 |
ENSRNOT00000009825
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 |
| chr14_+_66598259 | 26.56 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr4_+_136512201 | 26.51 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chr4_+_113970079 | 26.47 |
ENSRNOT00000079547
|
Rtkn
|
rhotekin |
| chr1_-_140262452 | 26.47 |
ENSRNOT00000046849
ENSRNOT00000045165 ENSRNOT00000025536 ENSRNOT00000041839 |
Ntrk3
|
neurotrophic receptor tyrosine kinase 3 |
| chr20_+_40236437 | 26.15 |
ENSRNOT00000000743
|
Hs3st5
|
heparan sulfate-glucosamine 3-sulfotransferase 5 |
| chr2_+_155718359 | 26.03 |
ENSRNOT00000077973
|
Kcnab1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr10_+_11393103 | 25.85 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr1_+_220322940 | 25.80 |
ENSRNOT00000074972
|
LOC108348085
|
beta-1,4-glucuronyltransferase 1 |
| chr14_+_12218553 | 25.75 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr15_+_34251606 | 25.64 |
ENSRNOT00000025725
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr3_-_66885085 | 25.50 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr19_-_58735173 | 25.47 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr10_-_45534570 | 25.34 |
ENSRNOT00000058362
|
Gjc2
|
gap junction protein, gamma 2 |
| chr2_+_54191538 | 25.32 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_+_145257714 | 25.29 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr11_-_87449940 | 25.19 |
ENSRNOT00000002560
|
Slc7a4
|
solute carrier family 7, member 4 |
| chr8_+_119030875 | 25.06 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr20_+_2004052 | 24.95 |
ENSRNOT00000001008
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr4_+_71675383 | 24.92 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr13_-_76049363 | 24.33 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr5_-_40237591 | 24.20 |
ENSRNOT00000011393
|
Fut9
|
fucosyltransferase 9 |
| chr3_+_80075991 | 23.87 |
ENSRNOT00000080266
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr15_+_33600102 | 23.80 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chrX_+_69730242 | 23.44 |
ENSRNOT00000075980
ENSRNOT00000076425 |
Eda
|
ectodysplasin-A |
| chr15_+_4240203 | 23.39 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr12_+_49761120 | 23.18 |
ENSRNOT00000070961
|
Myo18b
|
myosin XVIIIb |
| chr10_+_90930010 | 23.15 |
ENSRNOT00000003765
|
Higd1b
|
HIG1 hypoxia inducible domain family, member 1B |
| chrX_+_92131209 | 23.04 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr7_-_121029754 | 23.00 |
ENSRNOT00000004703
|
Nptxr
|
neuronal pentraxin receptor |
| chr1_+_262892545 | 22.81 |
ENSRNOT00000068278
ENSRNOT00000024709 ENSRNOT00000024750 |
Kcnip2
|
potassium voltage-gated channel interacting protein 2 |
| chr3_+_56861396 | 22.75 |
ENSRNOT00000000008
ENSRNOT00000084375 |
Gad1
|
glutamate decarboxylase 1 |
| chr15_+_33600337 | 22.68 |
ENSRNOT00000075965
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr12_-_5773036 | 22.53 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr4_+_41364441 | 22.52 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr1_-_215423099 | 22.42 |
ENSRNOT00000027259
|
AABR07006049.1
|
|
| chr13_+_60435946 | 22.35 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr4_+_180291389 | 22.34 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr14_-_112946204 | 22.12 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr9_-_80166807 | 22.10 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr14_-_84106997 | 22.02 |
ENSRNOT00000065501
|
Osbp2
|
oxysterol binding protein 2 |
| chr18_-_38088457 | 22.01 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chrX_+_71342775 | 21.89 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr10_-_110101872 | 21.85 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr8_+_116715755 | 21.64 |
ENSRNOT00000090239
|
Camkv
|
CaM kinase-like vesicle-associated |
| chr7_+_120153184 | 21.58 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr5_-_79874671 | 21.45 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr16_-_39719187 | 21.40 |
ENSRNOT00000092971
|
Gpm6a
|
glycoprotein m6a |
| chr10_-_12916784 | 21.33 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr16_+_46731403 | 21.19 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr18_-_14016713 | 21.13 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr16_+_22250470 | 21.11 |
ENSRNOT00000015799
|
Lzts1
|
leucine zipper tumor suppressor 1 |
| chr7_-_98098268 | 21.09 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr1_-_215373234 | 20.98 |
ENSRNOT00000034299
|
AABR07006055.1
|
|
| chr1_+_198210525 | 20.87 |
ENSRNOT00000026755
|
Ypel3
|
yippee-like 3 |
| chr10_-_47724499 | 20.78 |
ENSRNOT00000085011
|
Rnf112
|
ring finger protein 112 |
| chr1_+_164225934 | 20.78 |
ENSRNOT00000034389
|
Map6
|
microtubule-associated protein 6 |
| chr10_-_81942188 | 20.75 |
ENSRNOT00000003784
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr12_-_25638797 | 20.73 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr4_+_110699557 | 20.72 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr5_+_169181418 | 20.53 |
ENSRNOT00000004508
|
Klhl21
|
kelch-like family member 21 |
| chr5_+_22380334 | 20.45 |
ENSRNOT00000009744
|
Clvs1
|
clavesin 1 |
| chrX_-_107542510 | 20.41 |
ENSRNOT00000074140
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr10_-_89338739 | 20.35 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr17_-_88037034 | 20.26 |
ENSRNOT00000080193
ENSRNOT00000034907 |
Prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr6_+_58467254 | 20.18 |
ENSRNOT00000065396
|
Etv1
|
ets variant 1 |
| chr6_-_67085390 | 19.94 |
ENSRNOT00000083277
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr1_+_220446425 | 19.86 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chrX_-_157013443 | 19.80 |
ENSRNOT00000082711
|
Srpk3
|
SRSF protein kinase 3 |
| chr15_+_48674380 | 19.37 |
ENSRNOT00000018762
|
Fbxo16
|
F-box protein 16 |
| chr1_+_97632473 | 19.34 |
ENSRNOT00000023671
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr4_+_184019087 | 19.28 |
ENSRNOT00000055435
|
Bicd1
|
BICD cargo adaptor 1 |
| chr1_+_189665976 | 19.10 |
ENSRNOT00000033673
|
Lyrm1
|
LYR motif containing 1 |
| chr1_+_255185629 | 19.07 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr4_-_153373649 | 19.06 |
ENSRNOT00000016495
|
Atp6v1e1
|
ATPase H+ transporting V1 subunit E1 |
| chr5_+_151692108 | 19.06 |
ENSRNOT00000086144
|
Fam46b
|
family with sequence similarity 46, member B |
| chr14_+_36071376 | 19.02 |
ENSRNOT00000082183
|
Lnx1
|
ligand of numb-protein X 1 |
| chr7_+_11383116 | 19.02 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr11_-_88038518 | 18.95 |
ENSRNOT00000051991
|
Rimbp3
|
RIMS binding protein 3 |
| chrX_+_1297099 | 18.95 |
ENSRNOT00000013522
|
Elk1
|
ELK1, ETS transcription factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myog_Tcf12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 37.5 | 112.4 | GO:0055073 | cadmium ion homeostasis(GO:0055073) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 36.6 | 146.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 25.7 | 103.0 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 21.7 | 65.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 19.9 | 59.6 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 19.8 | 59.5 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 18.8 | 56.4 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 18.5 | 73.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 16.0 | 79.8 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 14.7 | 117.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 13.6 | 40.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 13.4 | 80.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 12.6 | 50.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 12.2 | 61.2 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) NMDA selective glutamate receptor signaling pathway(GO:0098989) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 12.0 | 59.8 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 10.8 | 86.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 10.7 | 32.0 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of synaptic vesicle recycling(GO:1903422) |
| 10.7 | 53.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 10.0 | 30.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 9.9 | 39.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 9.6 | 66.9 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 9.4 | 37.8 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 8.7 | 34.8 | GO:0021586 | pons maturation(GO:0021586) |
| 8.6 | 25.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 8.6 | 77.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 8.6 | 179.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 8.1 | 32.5 | GO:0031179 | peptide modification(GO:0031179) |
| 8.1 | 72.9 | GO:0036376 | sodium ion export from cell(GO:0036376) regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 8.0 | 64.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 7.9 | 47.6 | GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051343) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 7.8 | 23.4 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 7.7 | 31.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 7.6 | 22.8 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 7.4 | 29.7 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 7.2 | 28.6 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 6.9 | 34.3 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 6.8 | 20.3 | GO:0006178 | guanine salvage(GO:0006178) GMP catabolic process(GO:0046038) guanine biosynthetic process(GO:0046099) |
| 6.7 | 33.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 6.6 | 26.5 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 6.4 | 38.6 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 6.4 | 38.3 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 6.2 | 18.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 5.6 | 22.5 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 5.6 | 39.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 5.6 | 16.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 5.5 | 22.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 5.5 | 27.5 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 5.5 | 16.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 5.4 | 59.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 5.4 | 21.4 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 5.3 | 42.2 | GO:0015871 | choline transport(GO:0015871) |
| 5.2 | 36.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 5.1 | 35.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 5.1 | 15.4 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 4.9 | 38.8 | GO:0035900 | response to isolation stress(GO:0035900) |
| 4.8 | 19.3 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 4.7 | 14.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 4.7 | 55.9 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 4.6 | 22.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 4.3 | 17.1 | GO:1904426 | positive regulation of GTP binding(GO:1904426) |
| 4.3 | 29.9 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 4.3 | 21.3 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 4.3 | 8.5 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 4.2 | 8.5 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 4.2 | 21.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 4.2 | 12.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 4.2 | 16.7 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 4.1 | 16.2 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 4.0 | 16.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 4.0 | 12.0 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 4.0 | 15.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 3.9 | 31.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 3.9 | 11.7 | GO:0061146 | ureter maturation(GO:0035799) Peyer's patch morphogenesis(GO:0061146) |
| 3.8 | 26.9 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 3.8 | 19.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 3.8 | 22.5 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 3.6 | 10.9 | GO:1904954 | regulation of collateral sprouting in absence of injury(GO:0048696) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 3.6 | 17.9 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 3.5 | 10.4 | GO:1904722 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 3.4 | 78.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 3.4 | 47.6 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 3.4 | 3.4 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 3.2 | 28.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 3.2 | 76.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 3.1 | 12.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 3.1 | 12.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 3.0 | 81.7 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 3.0 | 24.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 2.9 | 8.8 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 2.9 | 17.6 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 2.8 | 109.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 2.8 | 30.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 2.8 | 16.6 | GO:0046959 | habituation(GO:0046959) |
| 2.7 | 16.0 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 2.7 | 18.6 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 2.7 | 8.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 2.7 | 13.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 2.6 | 21.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 2.6 | 41.7 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 2.5 | 24.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 2.5 | 9.9 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 2.4 | 9.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 2.4 | 9.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 2.4 | 35.7 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 2.4 | 26.0 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 2.3 | 14.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 2.3 | 16.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 2.3 | 6.9 | GO:0015966 | MAPK import into nucleus(GO:0000189) diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 2.2 | 2.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 2.2 | 11.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 2.2 | 28.8 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 2.2 | 39.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 2.1 | 14.9 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 2.1 | 25.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 2.1 | 6.4 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.1 | 8.5 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 2.1 | 18.7 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 2.1 | 12.4 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 2.0 | 30.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 2.0 | 8.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 2.0 | 56.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 2.0 | 6.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 2.0 | 11.8 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 2.0 | 7.8 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 2.0 | 13.7 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 2.0 | 9.8 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 1.9 | 119.3 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 1.9 | 28.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 1.9 | 14.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 1.8 | 9.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 1.8 | 25.8 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 1.8 | 5.4 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 1.8 | 3.5 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 1.7 | 6.9 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 1.7 | 24.2 | GO:0036065 | fucosylation(GO:0036065) |
| 1.7 | 3.5 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 1.7 | 22.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 1.7 | 10.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 1.7 | 5.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.7 | 5.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.6 | 44.2 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 1.6 | 14.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 1.6 | 32.5 | GO:0014741 | negative regulation of muscle hypertrophy(GO:0014741) |
| 1.6 | 12.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 1.5 | 10.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 1.5 | 38.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 1.5 | 4.6 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 1.5 | 6.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) oxidative single-stranded RNA demethylation(GO:0035553) |
| 1.5 | 22.2 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 1.5 | 5.9 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 1.5 | 19.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.4 | 2.9 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 1.4 | 4.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 1.4 | 7.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 1.4 | 4.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.4 | 25.9 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 1.4 | 67.9 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 1.4 | 14.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.3 | 17.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 1.3 | 26.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 1.3 | 2.6 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 1.3 | 16.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 1.3 | 6.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.3 | 10.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 1.3 | 33.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 1.3 | 6.3 | GO:0055119 | relaxation of cardiac muscle(GO:0055119) regulation of relaxation of cardiac muscle(GO:1901897) |
| 1.3 | 8.8 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 1.2 | 23.7 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 1.2 | 27.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 1.2 | 31.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 1.2 | 14.8 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.2 | 6.2 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 1.2 | 7.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 1.2 | 21.8 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 1.2 | 17.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 1.2 | 15.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 1.2 | 24.9 | GO:0051593 | response to folic acid(GO:0051593) |
| 1.2 | 4.7 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.2 | 10.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 1.1 | 6.8 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.1 | 20.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 1.1 | 16.8 | GO:0032095 | regulation of response to food(GO:0032095) |
| 1.0 | 12.6 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 1.0 | 10.4 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.0 | 14.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 1.0 | 9.2 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 1.0 | 3.0 | GO:1990402 | embryonic liver development(GO:1990402) |
| 1.0 | 34.3 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 1.0 | 10.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 1.0 | 44.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 1.0 | 2.9 | GO:0051142 | positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 1.0 | 1.9 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.9 | 6.6 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.9 | 2.8 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.9 | 55.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.9 | 9.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.9 | 14.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.9 | 7.0 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.9 | 13.9 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.9 | 18.9 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.8 | 5.9 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.8 | 5.8 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.8 | 9.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.8 | 11.3 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.8 | 4.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.8 | 12.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.8 | 10.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.8 | 17.6 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.8 | 38.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.8 | 8.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.7 | 2.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.7 | 11.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.7 | 2.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.7 | 34.0 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.7 | 5.8 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.7 | 17.7 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.7 | 24.6 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.7 | 7.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.7 | 34.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.7 | 5.6 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.7 | 5.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.7 | 16.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.7 | 10.7 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.7 | 36.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.7 | 72.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.7 | 11.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.7 | 2.0 | GO:0034034 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.7 | 7.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.6 | 5.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.6 | 3.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.6 | 2.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 9.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.6 | 45.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.6 | 0.6 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.6 | 26.5 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.6 | 11.7 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.5 | 16.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 4.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.5 | 40.9 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.5 | 3.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.5 | 2.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.5 | 9.2 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.5 | 1.9 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.5 | 13.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.5 | 3.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.5 | 6.0 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.5 | 5.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.5 | 2.3 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.5 | 9.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.4 | 5.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.4 | 7.4 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.4 | 15.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.4 | 54.4 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.4 | 0.8 | GO:0050968 | detection of chemical stimulus involved in sensory perception of pain(GO:0050968) regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.4 | 6.7 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.4 | 10.1 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.4 | 5.4 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.4 | 1.6 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.4 | 15.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.4 | 10.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.4 | 4.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 8.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 36.1 | GO:0050808 | synapse organization(GO:0050808) |
| 0.4 | 4.6 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) |
| 0.4 | 2.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.4 | 8.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.4 | 1.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 19.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.3 | 1.8 | GO:2001053 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.3 | 2.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 7.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.3 | 8.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 4.6 | GO:0036475 | neuron death in response to oxidative stress(GO:0036475) |
| 0.3 | 11.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 14.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.3 | 4.3 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.3 | 17.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.2 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 9.0 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.2 | 2.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 0.6 | GO:1903925 | hormone-mediated apoptotic signaling pathway(GO:0008628) cellular response to magnetism(GO:0071259) response to bisphenol A(GO:1903925) |
| 0.2 | 1.5 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.2 | 6.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.2 | 2.7 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.2 | 18.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.2 | 3.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.2 | 0.6 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.2 | 11.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 1.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 15.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.2 | 5.9 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 7.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.2 | 16.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.2 | 3.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 | 4.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 2.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.2 | 1.7 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.2 | 2.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 2.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 19.9 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.1 | 0.6 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.4 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 2.8 | GO:0009067 | aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.1 | 0.7 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.6 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.7 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 4.0 | GO:0008306 | associative learning(GO:0008306) |
| 0.1 | 0.8 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 0.4 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.1 | 2.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.4 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 2.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 61.2 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 14.1 | 112.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 14.0 | 55.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 13.1 | 39.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 13.0 | 39.0 | GO:0043511 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin complex(GO:0043511) |
| 12.7 | 38.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 12.2 | 72.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 11.6 | 34.8 | GO:0098855 | HCN channel complex(GO:0098855) |
| 11.6 | 34.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 11.4 | 136.8 | GO:0005861 | troponin complex(GO:0005861) |
| 11.2 | 56.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 9.5 | 47.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 9.1 | 54.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 8.7 | 52.0 | GO:1990393 | 3M complex(GO:1990393) |
| 8.6 | 34.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 6.8 | 27.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 6.4 | 19.3 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 6.1 | 122.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 5.8 | 17.5 | GO:0070821 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 5.8 | 298.1 | GO:0031672 | A band(GO:0031672) |
| 5.7 | 17.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 5.2 | 62.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 5.1 | 30.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 4.9 | 63.5 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 4.5 | 50.0 | GO:0032009 | early phagosome(GO:0032009) |
| 4.4 | 8.9 | GO:0044305 | calyx of Held(GO:0044305) |
| 3.5 | 101.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 3.3 | 9.8 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.9 | 20.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 2.8 | 8.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 2.8 | 13.8 | GO:0070695 | FHF complex(GO:0070695) |
| 2.6 | 85.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 2.6 | 23.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 2.5 | 10.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 2.5 | 29.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 2.5 | 29.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 2.5 | 32.0 | GO:0016342 | catenin complex(GO:0016342) |
| 2.4 | 7.3 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 2.4 | 44.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 2.4 | 244.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 2.3 | 16.2 | GO:0008091 | spectrin(GO:0008091) |
| 2.2 | 17.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 2.2 | 11.0 | GO:0097361 | CIA complex(GO:0097361) |
| 2.2 | 17.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 2.1 | 8.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 2.1 | 56.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 2.1 | 34.9 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 1.9 | 239.4 | GO:0030017 | sarcomere(GO:0030017) |
| 1.8 | 16.2 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 1.7 | 44.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.7 | 19.0 | GO:0002177 | manchette(GO:0002177) |
| 1.6 | 9.5 | GO:0071547 | piP-body(GO:0071547) |
| 1.6 | 25.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.5 | 44.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.5 | 47.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.5 | 7.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.4 | 31.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.3 | 8.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 1.3 | 5.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 1.3 | 3.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.3 | 8.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.2 | 18.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 1.2 | 18.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.1 | 7.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.1 | 18.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 31.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.0 | 11.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.0 | 70.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 1.0 | 3.0 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 1.0 | 193.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.9 | 6.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.9 | 32.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.9 | 15.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.9 | 14.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.8 | 231.0 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.8 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.8 | 9.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 5.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.8 | 2.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.8 | 12.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.8 | 19.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.8 | 6.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.8 | 4.6 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.8 | 65.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.8 | 40.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.7 | 17.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.7 | 72.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.7 | 11.2 | GO:0043034 | costamere(GO:0043034) |
| 0.7 | 3.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 14.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.6 | 6.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.6 | 2.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 29.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.6 | 7.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.5 | 4.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.5 | 10.9 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.5 | 4.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.5 | 89.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.5 | 12.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.5 | 30.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.5 | 10.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.5 | 11.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 4.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.4 | 8.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.4 | 4.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.4 | 42.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.4 | 18.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.4 | 39.0 | GO:0016605 | PML body(GO:0016605) |
| 0.4 | 86.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.4 | 22.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.4 | 12.4 | GO:0005844 | polysome(GO:0005844) |
| 0.4 | 9.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 16.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 2.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 3.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 3.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 9.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 12.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.3 | 7.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.3 | 2.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 50.2 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.3 | 5.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 1.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.3 | 11.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.3 | 8.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 6.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 2.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.2 | 7.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 39.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 43.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 9.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 3.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 11.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 5.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 8.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 16.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 1.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 6.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.0 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 3.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.0 | 66.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 19.8 | 59.5 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 16.0 | 79.8 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 15.9 | 47.6 | GO:0031997 | N-terminal myristoylation domain binding(GO:0031997) calcium ion binding involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099567) |
| 15.5 | 15.5 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 14.0 | 55.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 12.4 | 86.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 12.3 | 73.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 11.0 | 55.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 10.9 | 65.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 10.3 | 71.8 | GO:0034711 | inhibin binding(GO:0034711) |
| 10.2 | 40.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 10.2 | 112.4 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 9.8 | 49.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 9.5 | 47.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 9.3 | 74.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 9.1 | 45.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 8.8 | 61.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 8.4 | 33.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 8.3 | 115.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 8.1 | 32.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 7.7 | 23.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 7.4 | 73.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 7.2 | 35.8 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 7.1 | 42.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 6.9 | 20.8 | GO:2001070 | starch binding(GO:2001070) |
| 6.8 | 20.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 6.5 | 78.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 6.4 | 19.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 6.4 | 38.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 6.4 | 25.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 6.0 | 54.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 5.9 | 77.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 5.7 | 39.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 5.4 | 59.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 5.2 | 36.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 5.2 | 72.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 5.1 | 25.7 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 4.9 | 59.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 4.9 | 39.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 4.8 | 28.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 4.7 | 61.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 4.7 | 285.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 4.6 | 22.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 4.3 | 12.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 4.3 | 163.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 4.1 | 101.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 4.0 | 24.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 3.6 | 25.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 3.6 | 21.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 3.5 | 28.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 3.5 | 34.8 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 3.5 | 10.4 | GO:0031249 | denatured protein binding(GO:0031249) |
| 3.3 | 10.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 3.2 | 32.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 3.2 | 99.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 3.1 | 12.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 3.0 | 26.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 2.9 | 8.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 2.8 | 16.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 2.8 | 14.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 2.8 | 8.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 2.8 | 80.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 2.7 | 16.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 2.7 | 21.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 2.6 | 26.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 2.6 | 15.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 2.4 | 34.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 2.2 | 42.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 2.2 | 13.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 2.1 | 30.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 2.1 | 17.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 2.1 | 100.5 | GO:0019213 | deacetylase activity(GO:0019213) |
| 2.1 | 6.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 2.1 | 65.6 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 2.1 | 14.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 2.1 | 51.5 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 2.1 | 10.3 | GO:0035473 | lipase binding(GO:0035473) |
| 1.9 | 5.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.9 | 24.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.9 | 45.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.9 | 39.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 1.9 | 13.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 1.9 | 5.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.8 | 73.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 1.8 | 12.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.7 | 3.5 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 1.7 | 10.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.7 | 5.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.6 | 12.9 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 1.6 | 34.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.6 | 12.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 1.5 | 12.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.5 | 16.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.5 | 6.1 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 1.5 | 18.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 1.5 | 33.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.5 | 14.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.5 | 7.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.5 | 36.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 1.4 | 5.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 1.4 | 33.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 1.4 | 6.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 1.2 | 29.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 1.2 | 31.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.2 | 34.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 1.2 | 24.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 1.1 | 31.9 | GO:0005501 | retinoid binding(GO:0005501) |
| 1.1 | 71.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 1.1 | 35.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 1.1 | 29.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.1 | 28.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.0 | 10.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 1.0 | 14.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.0 | 9.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.0 | 58.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 1.0 | 5.9 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.0 | 41.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.9 | 29.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.9 | 174.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.9 | 20.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.9 | 14.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.9 | 29.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.9 | 10.3 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.8 | 19.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.8 | 2.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.7 | 12.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.6 | 18.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 10.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.6 | 1.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.6 | 2.4 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.6 | 17.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.6 | 2.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.6 | 6.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.6 | 20.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.5 | 47.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.5 | 1.6 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.5 | 4.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 44.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.5 | 33.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.5 | 21.5 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.5 | 3.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.5 | 8.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 23.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 3.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.5 | 2.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 9.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 11.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 0.8 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.4 | 11.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 16.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.4 | 3.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 1.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 10.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.3 | 21.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.3 | 35.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 1.9 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 4.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 31.1 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.3 | 94.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.3 | 4.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.3 | 2.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 14.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 27.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.2 | 2.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 3.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 80.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.2 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 12.7 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.2 | 15.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.2 | 9.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 27.8 | GO:0020037 | heme binding(GO:0020037) |
| 0.2 | 13.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.2 | 0.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 2.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 75.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.2 | 0.6 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 3.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 78.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 7.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 5.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 3.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 3.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 3.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.0 | GO:0016893 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters(GO:0016893) |
| 0.1 | 2.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 6.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 20.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 5.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 147.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 2.1 | 35.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 2.0 | 45.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 1.9 | 66.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 1.9 | 39.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.6 | 54.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 1.6 | 28.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 1.3 | 37.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 1.1 | 12.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.1 | 63.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 1.1 | 15.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 1.0 | 16.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 1.0 | 34.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 1.0 | 21.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.8 | 36.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.8 | 33.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.8 | 37.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.7 | 34.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.6 | 25.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.6 | 29.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.6 | 10.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.6 | 29.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.6 | 39.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.6 | 10.6 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.5 | 7.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 5.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.5 | 18.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.5 | 89.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.4 | 132.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.4 | 3.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.4 | 10.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.4 | 11.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 15.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.4 | 11.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.4 | 10.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.4 | 10.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 19.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.3 | 7.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 3.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 5.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 3.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 8.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 6.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 6.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 11.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 2.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 4.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 165.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 8.3 | 314.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 4.1 | 143.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 3.9 | 39.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 3.6 | 54.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 3.4 | 47.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 3.2 | 78.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 3.1 | 28.0 | REACTOME DEFENSINS | Genes involved in Defensins |
| 3.0 | 76.9 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 2.9 | 25.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 2.6 | 97.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 2.1 | 54.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 1.8 | 23.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.8 | 25.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.7 | 30.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.7 | 35.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.6 | 16.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 1.6 | 43.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 1.5 | 22.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 1.5 | 18.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.4 | 38.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 1.3 | 64.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 1.3 | 102.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 1.2 | 17.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.2 | 17.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 1.2 | 15.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 1.2 | 17.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 1.1 | 20.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.1 | 5.5 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 1.0 | 11.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.9 | 16.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.9 | 15.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.8 | 29.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.8 | 16.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.8 | 12.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.8 | 48.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.8 | 58.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.7 | 5.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.7 | 10.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.6 | 12.9 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.6 | 10.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.5 | 11.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 19.7 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.5 | 10.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 2.9 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.4 | 5.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 8.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 7.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 3.5 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.4 | 5.7 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.3 | 3.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 5.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 14.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.3 | 2.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 10.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.3 | 3.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 3.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 2.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 3.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 4.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 11.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 13.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 11.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 7.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 6.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 2.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 2.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 8.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 5.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 2.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 10.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.6 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.1 | 7.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 5.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |