Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
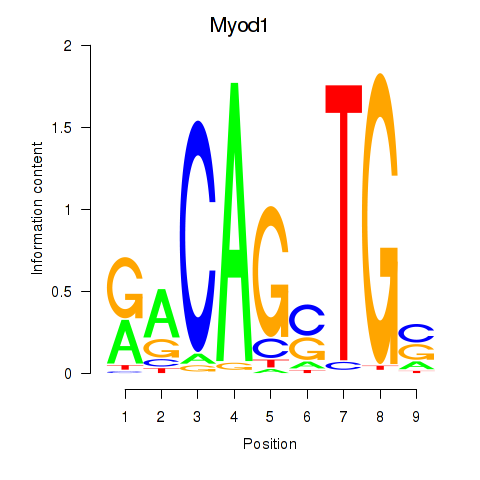
Results for Myod1
Z-value: 1.53
Transcription factors associated with Myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myod1
|
ENSRNOG00000011306 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myod1 | rn6_v1_chr1_+_102396538_102396538 | 0.65 | 2.5e-39 | Click! |
Activity profile of Myod1 motif
Sorted Z-values of Myod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_80321585 | 128.20 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr3_-_72219246 | 121.12 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr1_+_198655742 | 108.42 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr1_+_215609036 | 100.95 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr5_-_59025631 | 97.23 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr3_+_148386189 | 96.11 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr10_+_70262361 | 95.64 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr1_-_80331626 | 89.65 |
ENSRNOT00000022577
|
AABR07002677.1
|
|
| chr15_+_34251606 | 86.39 |
ENSRNOT00000025725
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr8_+_130416355 | 81.92 |
ENSRNOT00000026234
|
Klhl40
|
kelch-like family member 40 |
| chr9_+_14529218 | 81.38 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr9_+_82556573 | 77.64 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr10_-_94557764 | 72.34 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chr1_+_215609645 | 71.90 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr4_-_129619142 | 71.12 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chrX_-_111942749 | 70.55 |
ENSRNOT00000087583
|
AABR07040840.1
|
|
| chr10_-_95934345 | 69.34 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr13_+_52889737 | 60.80 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr16_-_40025401 | 58.58 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr1_-_213650247 | 56.58 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr15_+_2526368 | 52.60 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chr10_-_109729019 | 51.00 |
ENSRNOT00000054959
|
Ppp1r27
|
protein phosphatase 1, regulatory subunit 27 |
| chr9_-_115850634 | 48.87 |
ENSRNOT00000049720
|
Lrrc30
|
leucine rich repeat containing 30 |
| chr2_-_219842986 | 47.79 |
ENSRNOT00000055735
|
Agl
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr9_+_94286550 | 45.43 |
ENSRNOT00000026504
|
Chrnd
|
cholinergic receptor nicotinic delta subunit |
| chr7_+_11383116 | 40.41 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr1_+_266952561 | 39.99 |
ENSRNOT00000076452
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr20_-_5754773 | 37.41 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr2_+_11658568 | 35.41 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr1_-_264975132 | 34.98 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr6_-_99783047 | 34.30 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr10_+_92628356 | 32.82 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr8_-_33017854 | 31.46 |
ENSRNOT00000011386
|
Barx2
|
BARX homeobox 2 |
| chr15_+_4240203 | 29.89 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr3_+_79860179 | 29.80 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr2_+_189423559 | 29.37 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr10_-_8654892 | 27.93 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr4_-_99125111 | 26.38 |
ENSRNOT00000009184
|
Smyd1
|
SET and MYND domain containing 1 |
| chr10_+_34185898 | 25.69 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr10_-_89338739 | 25.60 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr5_+_61657507 | 24.79 |
ENSRNOT00000013228
|
Tmod1
|
tropomodulin 1 |
| chr10_-_62699723 | 24.70 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr8_+_65733400 | 24.26 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr3_-_117766120 | 23.19 |
ENSRNOT00000056022
|
Fbn1
|
fibrillin 1 |
| chr16_+_6962722 | 23.14 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr1_-_215846911 | 22.48 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr7_-_98098268 | 21.55 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr10_-_57243435 | 21.40 |
ENSRNOT00000005050
|
Chrne
|
cholinergic receptor nicotinic epsilon subunit |
| chr10_+_106812739 | 21.24 |
ENSRNOT00000074225
|
Syngr2
|
synaptogyrin 2 |
| chr19_-_37427989 | 21.07 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr4_+_57952982 | 19.95 |
ENSRNOT00000014465
|
Cpa1
|
carboxypeptidase A1 |
| chr11_+_27208564 | 19.78 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr1_-_31055453 | 19.41 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr1_-_128695995 | 19.12 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr3_-_110556808 | 19.11 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr8_+_32018560 | 18.56 |
ENSRNOT00000007358
|
Adamts8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr19_+_52032886 | 16.37 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr8_-_87419564 | 15.77 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr5_+_28850950 | 15.39 |
ENSRNOT00000009759
|
Tmem64
|
transmembrane protein 64 |
| chr5_+_75392790 | 15.12 |
ENSRNOT00000048457
|
Musk
|
muscle associated receptor tyrosine kinase |
| chr7_+_120153184 | 15.03 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr13_+_83681322 | 15.00 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr1_+_101884276 | 14.88 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr3_-_5617225 | 14.25 |
ENSRNOT00000008327
|
Tmem8c
|
transmembrane protein 8C |
| chr9_-_82419288 | 14.09 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr3_-_94418711 | 13.82 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr5_+_147476221 | 13.68 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr4_-_60358562 | 13.49 |
ENSRNOT00000018001
|
Chchd3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr3_-_123723524 | 12.99 |
ENSRNOT00000080351
|
Cenpb
|
centromere protein B |
| chr2_-_210782746 | 12.62 |
ENSRNOT00000025939
ENSRNOT00000081501 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr10_+_59173268 | 12.55 |
ENSRNOT00000013486
|
Ube2g1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr10_-_87517199 | 11.49 |
ENSRNOT00000080171
|
LOC100365588
|
keratin associated protein 1-3-like |
| chr10_+_83231409 | 11.44 |
ENSRNOT00000006230
|
Spop
|
speckle type BTB/POZ protein |
| chr20_-_45259928 | 11.33 |
ENSRNOT00000087226
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr10_+_83231238 | 11.03 |
ENSRNOT00000077625
|
Spop
|
speckle type BTB/POZ protein |
| chr1_+_72420352 | 10.92 |
ENSRNOT00000066307
|
Sbk3
|
SH3 domain binding kinase family, member 3 |
| chr1_+_100891866 | 10.55 |
ENSRNOT00000086890
ENSRNOT00000083336 |
Fuz
|
fuzzy planar cell polarity protein |
| chr10_+_36098051 | 10.33 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr5_+_3955227 | 10.16 |
ENSRNOT00000009908
|
Msc
|
musculin |
| chr1_-_80716143 | 10.14 |
ENSRNOT00000092048
ENSRNOT00000025829 |
Cblc
|
Cbl proto-oncogene C |
| chr1_+_220446425 | 10.07 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chr6_-_122721496 | 9.87 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chrX_-_111191932 | 9.84 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr5_-_144274981 | 9.26 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr1_-_265298797 | 9.02 |
ENSRNOT00000023137
|
Poll
|
DNA polymerase lambda |
| chr10_+_87788458 | 8.94 |
ENSRNOT00000042020
|
LOC100910814
|
keratin-associated protein 9-1-like |
| chr1_+_220353356 | 8.87 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr1_+_167374182 | 8.79 |
ENSRNOT00000092882
|
Stim1
|
stromal interaction molecule 1 |
| chrX_+_136466779 | 8.69 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chr10_-_87510629 | 8.68 |
ENSRNOT00000084197
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr1_+_85213652 | 8.48 |
ENSRNOT00000092044
|
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr10_-_87578854 | 8.35 |
ENSRNOT00000065619
|
LOC680160
|
similar to keratin associated protein 4-7 |
| chr7_+_2504695 | 7.91 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr6_-_72462657 | 7.90 |
ENSRNOT00000088720
ENSRNOT00000077578 |
Strn3
|
striatin 3 |
| chr10_-_87564327 | 7.83 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr9_-_82382272 | 7.82 |
ENSRNOT00000025627
|
Abcb6
|
ATP-binding cassette, subfamily B (MDR/TAP), member 6 |
| chr11_-_80826505 | 7.72 |
ENSRNOT00000032383
|
Rtp1
|
receptor (chemosensory) transporter protein 1 |
| chr2_+_54466280 | 7.71 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr5_-_151536518 | 7.39 |
ENSRNOT00000011311
|
Wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr5_+_48313599 | 7.36 |
ENSRNOT00000081825
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr3_-_2456041 | 7.17 |
ENSRNOT00000062001
|
Rnf224
|
ring finger protein 224 |
| chr11_-_83926524 | 7.10 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr4_-_147893992 | 6.93 |
ENSRNOT00000032158
|
Plxnd1
|
plexin D1 |
| chr7_-_139223116 | 6.89 |
ENSRNOT00000086559
|
Endou
|
endonuclease, poly(U)-specific |
| chr1_+_199287895 | 6.76 |
ENSRNOT00000026224
|
Stx4
|
syntaxin 4 |
| chr7_-_71226150 | 6.76 |
ENSRNOT00000005875
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr8_-_108109801 | 6.73 |
ENSRNOT00000034277
|
Sox14
|
SRY box 14 |
| chr6_+_127207732 | 6.73 |
ENSRNOT00000071976
|
RGD1309028
|
similar to RIKEN cDNA A830059I20 |
| chr8_-_120446455 | 6.58 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr1_+_218487824 | 6.39 |
ENSRNOT00000017958
|
Mrgprd
|
MAS related GPR family member D |
| chr15_-_8931983 | 6.28 |
ENSRNOT00000085322
|
Thrb
|
thyroid hormone receptor beta |
| chr10_-_87753668 | 6.15 |
ENSRNOT00000066758
ENSRNOT00000047104 |
Krtap4-3
|
keratin associated protein 4-3 |
| chr4_-_132111079 | 6.01 |
ENSRNOT00000013719
|
Eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr20_+_22728208 | 5.93 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr1_-_88690534 | 5.90 |
ENSRNOT00000072570
|
Ovol3
|
ovo-like zinc finger 3 |
| chr5_-_113578928 | 5.88 |
ENSRNOT00000010621
|
Plaa
|
phospholipase A2, activating protein |
| chr7_-_97977135 | 5.86 |
ENSRNOT00000008785
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr18_-_59819113 | 5.62 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr1_+_219345918 | 5.49 |
ENSRNOT00000025018
|
Cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr19_+_22699808 | 5.40 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr3_-_72171078 | 5.27 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr13_-_73704678 | 5.15 |
ENSRNOT00000005280
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr5_-_159371313 | 5.09 |
ENSRNOT00000009921
|
Padi1
|
peptidyl arginine deiminase 1 |
| chr3_-_60460724 | 5.04 |
ENSRNOT00000024706
|
Chrna1
|
cholinergic receptor nicotinic alpha 1 subunit |
| chr1_+_199287710 | 5.04 |
ENSRNOT00000082121
|
Stx4
|
syntaxin 4 |
| chr13_-_73704480 | 4.86 |
ENSRNOT00000005296
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr2_+_148608056 | 4.84 |
ENSRNOT00000015119
|
Tsc22d2
|
TSC22 domain family, member 2 |
| chr10_+_87832743 | 4.78 |
ENSRNOT00000055286
|
Krtap31-1
|
keratin associated protein 31-1 |
| chr17_-_13393243 | 4.32 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr10_+_11393103 | 4.10 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr2_+_148722231 | 4.07 |
ENSRNOT00000018022
|
Eif2a
|
eukaryotic translation initiation factor 2A |
| chr10_-_87510360 | 3.88 |
ENSRNOT00000077126
|
Krtap1-3
|
keratin associated protein 1-3 |
| chr5_+_116963609 | 3.82 |
ENSRNOT00000072724
ENSRNOT00000073487 |
AABR07049320.1
|
|
| chr19_-_55510460 | 3.72 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr9_+_76440186 | 3.72 |
ENSRNOT00000049479
|
RGD1562431
|
similar to chromosome 20 open reading frame 81 |
| chr10_+_90731865 | 3.69 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr13_+_89718066 | 3.52 |
ENSRNOT00000005147
|
Dedd
|
death effector domain-containing |
| chr8_-_122987191 | 3.41 |
ENSRNOT00000033976
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr20_-_11546046 | 3.38 |
ENSRNOT00000068374
|
LOC690386
|
hypothetical protein LOC690386 |
| chr1_+_171188133 | 3.35 |
ENSRNOT00000026511
|
Olr231
|
olfactory receptor 231 |
| chr2_+_118746333 | 3.27 |
ENSRNOT00000079431
|
AABR07009965.1
|
|
| chr19_-_55300403 | 3.17 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr8_-_116469915 | 3.14 |
ENSRNOT00000024193
|
Sema3f
|
semaphorin 3F |
| chr16_+_20873642 | 3.06 |
ENSRNOT00000004891
|
Ddx49
|
DEAD-box helicase 49 |
| chr3_+_110508403 | 3.03 |
ENSRNOT00000079959
|
AABR07053500.2
|
|
| chr10_+_17327275 | 2.96 |
ENSRNOT00000005486
|
Ubtd2
|
ubiquitin domain containing 2 |
| chrX_+_111396995 | 2.79 |
ENSRNOT00000087634
|
Pih1d3
|
PIH1 domain containing 3 |
| chr16_-_20873344 | 2.78 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr1_-_22480522 | 2.77 |
ENSRNOT00000046379
|
Taar7e
|
trace-amine-associated receptor 7e |
| chr10_+_87846947 | 2.76 |
ENSRNOT00000077436
|
Krtap9-5
|
keratin associated protein 9-5 |
| chr10_+_87819150 | 2.63 |
ENSRNOT00000073764
ENSRNOT00000064626 |
LOC680442
|
hypothetical protein LOC680442 |
| chr9_+_69790831 | 2.61 |
ENSRNOT00000045156
|
LOC100360781
|
ribosomal protein L37-like |
| chr10_+_90731148 | 2.51 |
ENSRNOT00000093604
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr20_-_27083410 | 2.48 |
ENSRNOT00000000432
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chr15_-_108120279 | 2.48 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr10_+_87782376 | 2.44 |
ENSRNOT00000017415
|
LOC680396
|
hypothetical protein LOC680396 |
| chr11_+_44877859 | 2.40 |
ENSRNOT00000060838
|
Col8a1
|
collagen type VIII alpha 1 chain |
| chr9_+_66335492 | 2.39 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr4_+_87440996 | 2.37 |
ENSRNOT00000074184
|
Vom1r68
|
vomeronasal 1 receptor 68 |
| chr1_-_175895510 | 2.34 |
ENSRNOT00000064535
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr1_+_41325462 | 2.31 |
ENSRNOT00000081017
ENSRNOT00000078494 ENSRNOT00000088168 |
Esr1
|
estrogen receptor 1 |
| chr4_+_65637407 | 2.21 |
ENSRNOT00000047954
|
Trim24
|
tripartite motif-containing 24 |
| chr2_-_184289126 | 2.20 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr9_+_88357556 | 2.19 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
| chr4_-_100099517 | 2.18 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr18_+_57011575 | 2.10 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr2_+_58462949 | 2.08 |
ENSRNOT00000080618
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr2_-_94730978 | 2.02 |
ENSRNOT00000087598
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr11_-_45510961 | 1.97 |
ENSRNOT00000002238
|
Tomm70
|
translocase of outer mitochondrial membrane 70 |
| chr20_+_31313018 | 1.93 |
ENSRNOT00000036719
|
Tysnd1
|
trypsin domain containing 1 |
| chr3_-_119678092 | 1.82 |
ENSRNOT00000085562
|
Ciao1
|
cytosolic iron-sulfur assembly component 1 |
| chr1_-_80783898 | 1.79 |
ENSRNOT00000045306
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chrX_-_112328642 | 1.73 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr10_+_64336200 | 1.71 |
ENSRNOT00000046519
|
Rpl37
|
ribosomal protein L37 |
| chr1_-_22488131 | 1.60 |
ENSRNOT00000051416
|
Taar7d
|
trace-amine-associated receptor 7d |
| chr8_-_68525911 | 1.45 |
ENSRNOT00000080588
ENSRNOT00000011109 |
Iqch
|
IQ motif containing H |
| chr5_+_58937615 | 1.42 |
ENSRNOT00000080909
|
Tesk1
|
testis-specific kinase 1 |
| chr1_+_230030914 | 1.35 |
ENSRNOT00000075730
|
Olr354
|
olfactory receptor 354 |
| chr2_-_172361779 | 1.28 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr15_+_27438853 | 1.28 |
ENSRNOT00000011636
|
Olr1622
|
olfactory receptor 1622 |
| chr10_+_97940705 | 1.20 |
ENSRNOT00000071153
|
Prkar1a
|
protein kinase cAMP-dependent type 1 regulatory subunit alpha |
| chr1_-_91042230 | 1.19 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr3_-_122057632 | 1.03 |
ENSRNOT00000038748
|
RGD1566226
|
similar to hypothetical protein F830045P16 |
| chrX_-_26376467 | 1.03 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr8_+_85489553 | 1.01 |
ENSRNOT00000082158
|
Rpl12-ps1
|
ribosomal protein L12, pseudogene 1 |
| chr10_+_44271587 | 0.96 |
ENSRNOT00000047700
|
Trim58
|
tripartite motif-containing 58 |
| chr10_+_17542374 | 0.93 |
ENSRNOT00000064079
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr1_-_22508170 | 0.91 |
ENSRNOT00000050763
|
Taar7c
|
trace amine-associated receptor 7C |
| chr18_+_14756684 | 0.87 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_-_134560713 | 0.77 |
ENSRNOT00000006621
|
Yaf2
|
YY1 associated factor 2 |
| chr13_+_101181994 | 0.66 |
ENSRNOT00000052407
|
Susd4
|
sushi domain containing 4 |
| chr8_-_39734594 | 0.66 |
ENSRNOT00000076872
|
Slc37a2
|
solute carrier family 37 member 2 |
| chr1_-_84856309 | 0.61 |
ENSRNOT00000079567
|
AABR07002783.1
|
|
| chr10_+_4313100 | 0.51 |
ENSRNOT00000074487
|
LOC100911685
|
eukaryotic peptide chain release factor GTP-binding subunit ERF3B-like |
| chr3_+_31802999 | 0.46 |
ENSRNOT00000041305
|
AABR07051996.1
|
|
| chrX_+_86126157 | 0.34 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr2_-_231521052 | 0.29 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr6_-_128727374 | 0.28 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chrX_+_159505344 | 0.20 |
ENSRNOT00000001164
|
Brs3
|
bombesin receptor subtype 3 |
| chr8_-_39734214 | 0.12 |
ENSRNOT00000040106
|
Slc37a2
|
solute carrier family 37 member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 32.0 | 96.1 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 20.3 | 81.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 16.4 | 81.9 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 12.3 | 86.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 12.0 | 95.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 11.8 | 35.4 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 9.0 | 72.3 | GO:0015871 | choline transport(GO:0015871) |
| 8.7 | 60.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 7.7 | 23.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 6.4 | 19.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 5.8 | 23.1 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 5.5 | 16.4 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 5.4 | 21.6 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 5.3 | 244.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 5.0 | 15.1 | GO:2000541 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 5.0 | 15.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 4.7 | 14.2 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 4.7 | 56.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 3.9 | 11.8 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 3.8 | 15.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 3.5 | 10.6 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 3.4 | 10.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 3.2 | 22.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 3.0 | 15.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 2.9 | 35.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 2.9 | 77.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 2.7 | 29.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 2.7 | 40.0 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 2.6 | 7.7 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 2.5 | 22.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 2.4 | 47.8 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 2.3 | 11.3 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 2.2 | 133.3 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 2.2 | 69.6 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 2.1 | 113.8 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 2.0 | 114.8 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 2.0 | 7.9 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.6 | 27.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.6 | 6.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 1.5 | 9.0 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 1.4 | 95.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 1.4 | 37.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 1.4 | 42.1 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 1.3 | 40.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 1.3 | 31.5 | GO:0001502 | cartilage condensation(GO:0001502) |
| 1.2 | 7.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 1.2 | 6.9 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.1 | 3.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 1.1 | 26.9 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 1.0 | 3.1 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.0 | 26.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.9 | 10.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.9 | 8.8 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.9 | 3.5 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.8 | 13.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.7 | 8.9 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.7 | 5.9 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.7 | 2.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.7 | 4.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.7 | 102.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.6 | 2.3 | GO:1990375 | baculum development(GO:1990375) |
| 0.5 | 13.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.5 | 5.9 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.5 | 3.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.5 | 5.5 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.5 | 37.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.4 | 2.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.4 | 2.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 7.7 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.3 | 9.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.3 | 35.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.3 | 12.6 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.3 | 2.2 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.3 | 4.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 10.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 6.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.2 | 14.9 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 2.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 4.1 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.2 | 7.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.2 | 13.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.2 | 2.1 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 6.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.9 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 1.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 6.9 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.2 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.1 | 0.8 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 2.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.0 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 3.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 6.1 | GO:0042633 | molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 | 2.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 2.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 1.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 9.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 3.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 20.0 | GO:0006605 | protein targeting(GO:0006605) |
| 0.0 | 0.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.1 | 126.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 15.5 | 77.6 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 12.3 | 147.6 | GO:0005861 | troponin complex(GO:0005861) |
| 5.5 | 307.3 | GO:0031672 | A band(GO:0031672) |
| 4.9 | 34.3 | GO:0008091 | spectrin(GO:0008091) |
| 4.9 | 24.3 | GO:0043293 | apoptosome(GO:0043293) |
| 4.8 | 19.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 3.6 | 72.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 3.6 | 71.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 3.5 | 56.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 2.9 | 130.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 2.3 | 29.8 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 2.1 | 23.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.9 | 13.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.7 | 99.4 | GO:0016459 | myosin complex(GO:0016459) |
| 1.7 | 11.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 1.3 | 9.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 1.1 | 7.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.0 | 56.6 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.9 | 13.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.8 | 16.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.7 | 23.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.7 | 91.5 | GO:0030017 | sarcomere(GO:0030017) |
| 0.7 | 37.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.7 | 3.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 46.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.6 | 15.0 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.6 | 78.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.6 | 4.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 4.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.4 | 2.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 2.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 23.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.4 | 53.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 1.8 | GO:0097361 | CIA complex(GO:0097361) |
| 0.4 | 7.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 7.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 2.2 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.3 | 7.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 10.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 2.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 34.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 26.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 9.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 5.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 58.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.7 | 172.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 24.0 | 96.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 18.3 | 128.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 13.6 | 81.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 12.0 | 191.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 8.0 | 47.8 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 5.5 | 254.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 4.2 | 71.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 4.2 | 37.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 3.9 | 11.8 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 3.5 | 35.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 3.3 | 29.8 | GO:0043495 | protein anchor(GO:0043495) |
| 3.2 | 13.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 3.2 | 60.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 3.0 | 15.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 2.5 | 15.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 2.4 | 72.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 2.2 | 88.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 2.2 | 69.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 1.9 | 11.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 1.9 | 56.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.8 | 9.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.6 | 7.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.5 | 7.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 1.5 | 34.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 1.0 | 22.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.0 | 5.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.0 | 52.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.9 | 7.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.8 | 7.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.8 | 7.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.8 | 6.9 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.7 | 20.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 10.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.7 | 35.1 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.7 | 3.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.6 | 6.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 10.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 7.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.5 | 14.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 4.5 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.5 | 8.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 2.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 25.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 12.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.4 | 16.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 12.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 6.9 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 5.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 10.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 52.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.2 | 28.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 12.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 30.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 4.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 73.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 24.1 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.8 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 2.1 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 5.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 4.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.2 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 1.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 6.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 7.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 34.7 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.0 | 1.3 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 36.6 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 3.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 96.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.7 | 143.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 1.4 | 63.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.1 | 23.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.0 | 26.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.8 | 102.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.7 | 6.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.6 | 11.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 21.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.5 | 23.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.5 | 9.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 11.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 15.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 15.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 4.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 9.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 4.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 3.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 26.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 7.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 2.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.5 | 409.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 6.0 | 71.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 3.0 | 106.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 2.4 | 47.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.5 | 63.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 1.4 | 35.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.7 | 8.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.6 | 7.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.6 | 118.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.5 | 8.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 70.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 12.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 4.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 11.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 4.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 7.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.3 | 5.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 2.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 7.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 14.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 7.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 1.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 8.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |