Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
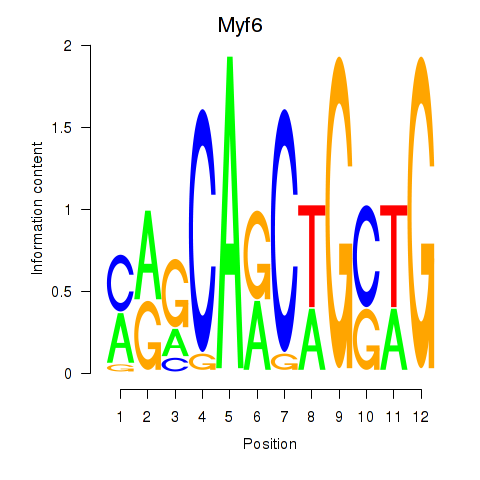
Results for Myf6
Z-value: 1.09
Transcription factors associated with Myf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myf6
|
ENSRNOG00000004878 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myf6 | rn6_v1_chr7_-_49741540_49741540 | -0.27 | 9.7e-07 | Click! |
Activity profile of Myf6 motif
Sorted Z-values of Myf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_31122093 | 32.47 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chr1_-_13915594 | 28.88 |
ENSRNOT00000015927
|
Arfgef3
|
ARFGEF family member 3 |
| chr15_+_50891127 | 23.76 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr17_+_88215834 | 23.27 |
ENSRNOT00000034098
|
Gpr158
|
G protein-coupled receptor 158 |
| chr3_+_71210301 | 21.94 |
ENSRNOT00000006504
|
Fam171b
|
family with sequence similarity 171, member B |
| chr5_+_144634143 | 20.99 |
ENSRNOT00000075558
|
RGD1563072
|
similar to hypothetical protein FLJ38984 |
| chr18_-_29015552 | 18.50 |
ENSRNOT00000028713
|
Nrg2
|
neuregulin 2 |
| chr10_-_90151042 | 18.45 |
ENSRNOT00000055187
|
Hdac5
|
histone deacetylase 5 |
| chr19_+_58823814 | 18.32 |
ENSRNOT00000027058
ENSRNOT00000088626 |
Kcnk1
|
potassium two pore domain channel subfamily K member 1 |
| chr8_+_49676540 | 18.13 |
ENSRNOT00000022032
ENSRNOT00000082205 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr1_-_252808380 | 17.18 |
ENSRNOT00000025856
|
Ch25h
|
cholesterol 25-hydroxylase |
| chr2_+_189906022 | 16.89 |
ENSRNOT00000016519
|
S100a13
|
S100 calcium binding protein A13 |
| chrX_-_15707436 | 16.28 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr8_-_103190243 | 16.27 |
ENSRNOT00000075305
|
Chst2
|
carbohydrate sulfotransferase 2 |
| chr4_-_64831473 | 16.12 |
ENSRNOT00000033268
|
Dgki
|
diacylglycerol kinase, iota |
| chr6_-_147172813 | 16.08 |
ENSRNOT00000066545
|
Itgb8
|
integrin subunit beta 8 |
| chr12_+_31530699 | 16.05 |
ENSRNOT00000033379
ENSRNOT00000067867 |
Rimbp2
|
RIMS binding protein 2 |
| chr5_+_98469047 | 15.62 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr7_+_54980120 | 15.55 |
ENSRNOT00000005690
ENSRNOT00000005773 |
Kcnc2
|
potassium voltage-gated channel subfamily C member 2 |
| chr19_+_10731855 | 15.46 |
ENSRNOT00000022277
|
Pllp
|
plasmolipin |
| chr2_+_149494330 | 15.23 |
ENSRNOT00000074764
ENSRNOT00000089865 |
Med12l
|
mediator complex subunit 12-like |
| chr4_-_64831233 | 15.21 |
ENSRNOT00000079285
|
Dgki
|
diacylglycerol kinase, iota |
| chr4_-_117490721 | 15.01 |
ENSRNOT00000021103
|
Nat8f3
|
N-acetyltransferase 8 (GCN5-related) family member 3 |
| chr6_+_64789940 | 14.32 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr9_-_82461903 | 14.09 |
ENSRNOT00000026654
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N |
| chr13_+_99173484 | 13.72 |
ENSRNOT00000080574
ENSRNOT00000088654 |
Lefty2
|
Left-right determination factor 2 |
| chr4_+_96562725 | 13.67 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr2_-_26699333 | 13.22 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr7_-_113937941 | 13.11 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr13_+_63526486 | 12.99 |
ENSRNOT00000003788
|
Brinp3
|
BMP/retinoic acid inducible neural specific 3 |
| chr5_-_82168347 | 12.92 |
ENSRNOT00000084959
ENSRNOT00000084147 |
Astn2
|
astrotactin 2 |
| chr6_-_27190126 | 12.37 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr4_-_64330996 | 12.27 |
ENSRNOT00000016088
|
Ptn
|
pleiotrophin |
| chr19_+_20607507 | 12.14 |
ENSRNOT00000000011
|
Cbln1
|
cerebellin 1 precursor |
| chr5_+_137371825 | 12.05 |
ENSRNOT00000072816
|
Tmem125
|
transmembrane protein 125 |
| chr6_+_132246602 | 12.03 |
ENSRNOT00000009896
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr9_+_9721105 | 11.97 |
ENSRNOT00000073042
ENSRNOT00000075494 |
C3
|
complement C3 |
| chr14_+_83752393 | 11.89 |
ENSRNOT00000081123
|
Selenom
|
selenoprotein M |
| chr5_+_148923098 | 11.75 |
ENSRNOT00000048781
|
Sdc3
|
syndecan 3 |
| chr18_+_83777665 | 11.66 |
ENSRNOT00000018682
|
Cbln2
|
cerebellin 2 precursor |
| chr9_+_118842787 | 11.49 |
ENSRNOT00000090512
|
Dlgap1
|
DLG associated protein 1 |
| chr20_-_4489281 | 11.31 |
ENSRNOT00000031548
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr18_-_38088457 | 11.31 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr4_+_181481147 | 11.29 |
ENSRNOT00000002523
|
Klhl42
|
kelch-like family, member 42 |
| chr3_+_41019898 | 10.99 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr3_-_44342355 | 10.71 |
ENSRNOT00000059280
|
Acvr1c
|
activin A receptor type 1C |
| chr14_-_83062302 | 10.47 |
ENSRNOT00000086769
ENSRNOT00000085735 |
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr14_+_75880410 | 10.42 |
ENSRNOT00000014078
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr1_-_64446818 | 10.35 |
ENSRNOT00000081980
|
Myadm
|
myeloid-associated differentiation marker |
| chr15_-_14622587 | 10.30 |
ENSRNOT00000086502
|
Synpr
|
synaptoporin |
| chr1_-_31055453 | 10.16 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr7_-_143793774 | 10.09 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr4_-_50860756 | 10.06 |
ENSRNOT00000068404
|
Cadps2
|
calcium dependent secretion activator 2 |
| chr12_+_48598647 | 9.95 |
ENSRNOT00000000889
|
Tmem119
|
transmembrane protein 119 |
| chr1_-_198454914 | 9.94 |
ENSRNOT00000049044
|
Prrt2
|
proline-rich transmembrane protein 2 |
| chr20_+_13067242 | 9.93 |
ENSRNOT00000083878
|
Dip2a
|
disco-interacting protein 2 homolog A |
| chr1_+_1702696 | 9.92 |
ENSRNOT00000019181
|
Lrp11
|
LDL receptor related protein 11 |
| chr2_+_23385183 | 9.85 |
ENSRNOT00000014860
|
Arsb
|
arylsulfatase B |
| chr9_+_16543688 | 9.84 |
ENSRNOT00000021868
|
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr12_-_42492526 | 9.74 |
ENSRNOT00000084018
|
Tbx3
|
T-box 3 |
| chr16_+_20691978 | 9.66 |
ENSRNOT00000038139
ENSRNOT00000082319 |
Tmem59l
|
transmembrane protein 59-like |
| chr10_+_102136283 | 9.60 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr13_-_113871842 | 9.58 |
ENSRNOT00000079549
|
Cr1l
|
complement C3b/C4b receptor 1 like |
| chr12_-_42492328 | 9.56 |
ENSRNOT00000011552
|
Tbx3
|
T-box 3 |
| chr4_-_113764532 | 9.54 |
ENSRNOT00000009269
|
Sema4f
|
ssemaphorin 4F |
| chr8_+_130401470 | 9.49 |
ENSRNOT00000043346
|
Zbtb47
|
zinc finger and BTB domain containing 47 |
| chr2_+_112868707 | 9.36 |
ENSRNOT00000017805
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr6_+_137997335 | 9.35 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr10_-_90995982 | 9.34 |
ENSRNOT00000093266
|
Gfap
|
glial fibrillary acidic protein |
| chr20_-_14545772 | 9.34 |
ENSRNOT00000001766
|
Bcr
|
BCR, RhoGEF and GTPase activating protein |
| chr2_-_231521052 | 9.32 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr4_-_39102807 | 9.28 |
ENSRNOT00000052063
|
Thsd7a
|
thrombospondin type 1 domain containing 7A |
| chr5_-_137372524 | 9.27 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chrX_-_104493714 | 9.11 |
ENSRNOT00000064458
ENSRNOT00000080386 |
Pcdh19
|
protocadherin 19 |
| chr1_+_91363492 | 9.04 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr4_+_157726941 | 9.03 |
ENSRNOT00000025081
|
Vamp1
|
vesicle-associated membrane protein 1 |
| chr11_+_31428358 | 9.00 |
ENSRNOT00000002827
|
Olig1
|
oligodendrocyte transcription factor 1 |
| chr13_-_39643361 | 8.95 |
ENSRNOT00000003527
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr8_-_28208466 | 8.92 |
ENSRNOT00000012247
|
Jam3
|
junctional adhesion molecule 3 |
| chr6_-_44361908 | 8.80 |
ENSRNOT00000009491
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr9_+_80118029 | 8.68 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr19_+_37476095 | 8.61 |
ENSRNOT00000092794
ENSRNOT00000023130 |
Hsd11b2
|
hydroxysteroid 11-beta dehydrogenase 2 |
| chrX_+_70596576 | 8.52 |
ENSRNOT00000045082
ENSRNOT00000003741 ENSRNOT00000076079 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr3_-_143063983 | 8.52 |
ENSRNOT00000006329
|
Napb
|
NSF attachment protein beta |
| chr3_-_9037942 | 8.49 |
ENSRNOT00000036770
|
Ier5l
|
immediate early response 5-like |
| chr5_-_77342299 | 8.34 |
ENSRNOT00000075994
|
RGD1566134
|
similar to alpha-2u-globulin |
| chr5_+_124690214 | 8.33 |
ENSRNOT00000011237
|
Plpp3
|
phospholipid phosphatase 3 |
| chr14_+_79538911 | 8.30 |
ENSRNOT00000009960
|
Sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr1_-_213973163 | 8.19 |
ENSRNOT00000024867
|
LOC100911402
|
cell cycle exit and neuronal differentiation protein 1-like |
| chr14_-_55081551 | 8.12 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr1_+_236031988 | 7.89 |
ENSRNOT00000016164
|
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr5_-_77408323 | 7.73 |
ENSRNOT00000046857
ENSRNOT00000046760 |
LOC259245
Mup4
|
alpha-2u globulin PGCL5 major urinary protein 4 |
| chr3_-_2534663 | 7.71 |
ENSRNOT00000049297
ENSRNOT00000044246 |
Grin1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr10_-_64642292 | 7.68 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr8_-_55144087 | 7.66 |
ENSRNOT00000039045
|
Dixdc1
|
DIX domain containing 1 |
| chr5_-_77492013 | 7.60 |
ENSRNOT00000012293
|
LOC259245
|
alpha-2u globulin PGCL5 |
| chr18_+_30004565 | 7.58 |
ENSRNOT00000027393
|
Pcdha4
|
protocadherin alpha 4 |
| chr4_+_32373641 | 7.56 |
ENSRNOT00000076086
|
Dlx6
|
distal-less homeobox 6 |
| chr4_-_62840357 | 7.55 |
ENSRNOT00000059892
|
Slc13a4
|
solute carrier family 13 member 4 |
| chr1_+_83965608 | 7.54 |
ENSRNOT00000079995
|
Cyp2t1
|
cytochrome P450, family 2, subfamily t, polypeptide 1 |
| chr8_+_408001 | 7.46 |
ENSRNOT00000046058
|
Gucy1a2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr1_+_213870502 | 7.44 |
ENSRNOT00000086483
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr1_-_265573117 | 7.43 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chrX_-_2657155 | 7.40 |
ENSRNOT00000005630
|
Chst7
|
carbohydrate sulfotransferase 7 |
| chrX_+_70596901 | 7.28 |
ENSRNOT00000088114
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr1_-_64030175 | 7.26 |
ENSRNOT00000089950
|
Tsen34l1
|
tRNA splicing endonuclease subunit 34-like 1 |
| chr4_-_88649216 | 7.24 |
ENSRNOT00000058626
|
Herc6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr9_-_55673704 | 7.21 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr15_+_104095179 | 7.12 |
ENSRNOT00000093487
|
Cldn10
|
claudin 10 |
| chr6_-_110904288 | 7.08 |
ENSRNOT00000014645
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr6_+_27768943 | 7.08 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr10_+_11393103 | 7.07 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr2_+_95008311 | 7.01 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chr4_+_113968995 | 6.97 |
ENSRNOT00000079511
|
Rtkn
|
rhotekin |
| chr7_+_145117951 | 6.96 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr2_+_95008477 | 6.95 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chr1_-_146029840 | 6.93 |
ENSRNOT00000016455
|
Mesdc1
|
mesoderm development candidate 1 |
| chr17_+_4846789 | 6.92 |
ENSRNOT00000073271
|
Gas1
|
growth arrest-specific 1 |
| chr3_-_2534375 | 6.91 |
ENSRNOT00000037725
|
Grin1
|
glutamate ionotropic receptor NMDA type subunit 1 |
| chr11_-_87449940 | 6.88 |
ENSRNOT00000002560
|
Slc7a4
|
solute carrier family 7, member 4 |
| chr5_-_17061837 | 6.87 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr16_+_56248331 | 6.82 |
ENSRNOT00000085300
|
Tusc3
|
tumor suppressor candidate 3 |
| chr11_+_33909439 | 6.74 |
ENSRNOT00000002310
|
Cbr3
|
carbonyl reductase 3 |
| chr10_+_58332588 | 6.71 |
ENSRNOT00000081954
|
Wscd1
|
WSC domain containing 1 |
| chr4_+_57019941 | 6.67 |
ENSRNOT00000011356
|
Smo
|
smoothened, frizzled class receptor |
| chr18_+_39172028 | 6.61 |
ENSRNOT00000086651
|
Kcnn2
|
potassium calcium-activated channel subfamily N member 2 |
| chr11_-_65759581 | 6.60 |
ENSRNOT00000034334
|
Lrrc58
|
leucine rich repeat containing 58 |
| chr4_+_157374318 | 6.60 |
ENSRNOT00000071027
|
AC115420.2
|
|
| chrX_-_156407404 | 6.59 |
ENSRNOT00000089814
|
Gdi1
|
GDP dissociation inhibitor 1 |
| chr7_-_58587787 | 6.51 |
ENSRNOT00000005814
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr3_+_161298962 | 6.50 |
ENSRNOT00000066028
|
Ctsa
|
cathepsin A |
| chr8_+_116332796 | 6.48 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr8_-_47404010 | 6.48 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr18_+_29972808 | 6.47 |
ENSRNOT00000074051
|
Pcdha4
|
protocadherin alpha 4 |
| chr7_+_59349745 | 6.45 |
ENSRNOT00000085334
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr8_+_48472824 | 6.37 |
ENSRNOT00000010463
ENSRNOT00000090780 |
Mcam
|
melanoma cell adhesion molecule |
| chr7_+_141326950 | 6.35 |
ENSRNOT00000084075
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr18_-_86878142 | 6.34 |
ENSRNOT00000058139
|
Dok6
|
docking protein 6 |
| chrX_-_84167717 | 6.32 |
ENSRNOT00000006415
|
Pof1b
|
premature ovarian failure 1B |
| chr2_+_3400977 | 6.31 |
ENSRNOT00000093593
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr1_-_170464341 | 6.30 |
ENSRNOT00000024850
|
Trim3
|
tripartite motif-containing 3 |
| chr16_-_69132584 | 6.29 |
ENSRNOT00000017776
|
Adgra2
|
adhesion G protein-coupled receptor A2 |
| chr8_+_118333706 | 6.26 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr11_+_33845463 | 6.25 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr17_+_13670520 | 6.22 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr10_-_82887301 | 6.16 |
ENSRNOT00000035894
|
Itga3
|
integrin subunit alpha 3 |
| chr11_-_70322690 | 6.14 |
ENSRNOT00000002443
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr17_+_81798756 | 6.07 |
ENSRNOT00000066826
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_+_89166890 | 5.96 |
ENSRNOT00000088331
|
Ramp2
|
receptor activity modifying protein 2 |
| chr5_-_77316764 | 5.94 |
ENSRNOT00000071395
ENSRNOT00000076464 |
Mup4
|
major urinary protein 4 |
| chr13_-_112099336 | 5.92 |
ENSRNOT00000009158
ENSRNOT00000044161 |
Camk1g
|
calcium/calmodulin-dependent protein kinase IG |
| chr1_+_262892545 | 5.90 |
ENSRNOT00000068278
ENSRNOT00000024709 ENSRNOT00000024750 |
Kcnip2
|
potassium voltage-gated channel interacting protein 2 |
| chr11_+_33812989 | 5.86 |
ENSRNOT00000042283
ENSRNOT00000075985 |
Cbr1
|
carbonyl reductase 1 |
| chr14_+_77380262 | 5.85 |
ENSRNOT00000008030
|
Nsg1
|
neuron specific gene family member 1 |
| chr9_-_19749145 | 5.84 |
ENSRNOT00000013956
|
Rcan2
|
regulator of calcineurin 2 |
| chr6_-_24563246 | 5.84 |
ENSRNOT00000074294
|
LOC685881
|
hypothetical protein LOC685881 |
| chr2_-_188672226 | 5.82 |
ENSRNOT00000027970
ENSRNOT00000050868 |
Adam15
|
ADAM metallopeptidase domain 15 |
| chrX_+_80213332 | 5.82 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr16_-_48437223 | 5.81 |
ENSRNOT00000013005
ENSRNOT00000059401 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr4_-_38240848 | 5.80 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr5_-_34813116 | 5.80 |
ENSRNOT00000017479
|
Nkain3
|
Sodium/potassium transporting ATPase interacting 3 |
| chr3_+_177225737 | 5.80 |
ENSRNOT00000045845
|
Oprl1
|
opioid related nociceptin receptor 1 |
| chr1_+_233382708 | 5.77 |
ENSRNOT00000019174
|
Gnaq
|
G protein subunit alpha q |
| chr8_+_75516904 | 5.70 |
ENSRNOT00000013142
|
Rora
|
RAR-related orphan receptor A |
| chr10_+_59799123 | 5.69 |
ENSRNOT00000026493
|
Trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr1_+_21525421 | 5.69 |
ENSRNOT00000017911
|
Arg1
|
arginase 1 |
| chr5_+_167141875 | 5.68 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr1_+_157573324 | 5.66 |
ENSRNOT00000092066
|
Rab30
|
RAB30, member RAS oncogene family |
| chr17_-_6244612 | 5.65 |
ENSRNOT00000042145
ENSRNOT00000090914 ENSRNOT00000082611 |
Ntrk2
|
neurotrophic receptor tyrosine kinase 2 |
| chr3_+_2648885 | 5.63 |
ENSRNOT00000020339
|
Abca2
|
ATP binding cassette subfamily A member 2 |
| chr1_-_255557055 | 5.62 |
ENSRNOT00000033780
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr3_-_134696654 | 5.58 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr5_-_130085838 | 5.57 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr4_+_114835064 | 5.57 |
ENSRNOT00000031964
|
LOC103692165
|
rhotekin |
| chr12_+_22165486 | 5.56 |
ENSRNOT00000001890
|
Mospd3
|
motile sperm domain containing 3 |
| chr1_+_101783621 | 5.52 |
ENSRNOT00000067679
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr2_-_18531210 | 5.52 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr8_+_36125999 | 5.51 |
ENSRNOT00000013036
ENSRNOT00000088046 |
Kirrel3
|
kin of IRRE like 3 (Drosophila) |
| chrX_+_69580015 | 5.51 |
ENSRNOT00000057920
|
Fam155b
|
family with sequence similarity 155, member B |
| chrX_+_92131209 | 5.50 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr1_-_47307488 | 5.50 |
ENSRNOT00000090033
|
Ezr
|
ezrin |
| chr20_+_31313018 | 5.48 |
ENSRNOT00000036719
|
Tysnd1
|
trypsin domain containing 1 |
| chr3_-_147143576 | 5.47 |
ENSRNOT00000091811
ENSRNOT00000012727 |
Snph
|
syntaphilin |
| chr1_+_220114228 | 5.43 |
ENSRNOT00000026718
|
Ctsf
|
cathepsin F |
| chr4_-_145147397 | 5.41 |
ENSRNOT00000010347
|
Lhfpl4
|
lipoma HMGIC fusion partner-like 4 |
| chr1_+_7305658 | 5.41 |
ENSRNOT00000056227
|
AABR07000261.1
|
|
| chr2_+_264786242 | 5.40 |
ENSRNOT00000015170
|
Ankrd13c
|
ankyrin repeat domain 13C |
| chr7_-_107009330 | 5.38 |
ENSRNOT00000074573
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr3_-_8924032 | 5.38 |
ENSRNOT00000023527
ENSRNOT00000085042 |
Sh3glb2
|
SH3 domain-containing GRB2-like endophilin B2 |
| chr3_+_175144495 | 5.34 |
ENSRNOT00000082601
ENSRNOT00000088026 |
Cdh4
|
cadherin 4 |
| chr10_+_89167146 | 5.33 |
ENSRNOT00000043953
|
Ramp2
|
receptor activity modifying protein 2 |
| chr7_+_70980422 | 5.29 |
ENSRNOT00000077912
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr10_+_90550147 | 5.28 |
ENSRNOT00000032944
|
Fzd2
|
frizzled class receptor 2 |
| chr1_-_80056574 | 5.27 |
ENSRNOT00000021200
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr3_-_122947075 | 5.24 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr7_-_70926903 | 5.24 |
ENSRNOT00000031005
|
Lrp1
|
LDL receptor related protein 1 |
| chr2_-_32518643 | 5.23 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_221801524 | 5.23 |
ENSRNOT00000031227
|
Nrxn2
|
neurexin 2 |
| chrX_-_157028434 | 5.22 |
ENSRNOT00000080088
|
Plxnb3
|
plexin B3 |
| chr17_-_86657473 | 5.20 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Myf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.3 | GO:0060932 | sinoatrial node cell development(GO:0060931) His-Purkinje system cell differentiation(GO:0060932) |
| 5.7 | 17.0 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 5.4 | 16.3 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 4.9 | 14.6 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 4.7 | 18.8 | GO:1901662 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 4.3 | 13.0 | GO:1904373 | response to kainic acid(GO:1904373) |
| 4.0 | 12.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 4.0 | 12.0 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 3.9 | 31.3 | GO:0046959 | habituation(GO:0046959) |
| 3.7 | 11.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 3.6 | 10.7 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 3.5 | 10.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 3.4 | 10.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 3.4 | 10.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 3.1 | 9.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 3.0 | 15.2 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 3.0 | 8.9 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 2.9 | 8.8 | GO:0003166 | bundle of His development(GO:0003166) |
| 2.6 | 15.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 2.6 | 7.9 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 2.6 | 18.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 2.5 | 7.5 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 2.5 | 7.4 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 2.4 | 16.9 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 2.3 | 9.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.3 | 6.9 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 2.3 | 29.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 2.2 | 6.7 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 2.2 | 4.4 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 2.1 | 8.5 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 2.1 | 6.2 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 2.0 | 5.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 1.9 | 9.5 | GO:0015755 | fructose transport(GO:0015755) |
| 1.9 | 17.1 | GO:0000050 | urea cycle(GO:0000050) |
| 1.9 | 3.7 | GO:0021586 | pons maturation(GO:0021586) |
| 1.8 | 18.4 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 1.8 | 11.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 1.8 | 20.0 | GO:0061042 | vascular wound healing(GO:0061042) |
| 1.8 | 10.6 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.8 | 5.3 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 1.8 | 10.5 | GO:0048143 | astrocyte activation(GO:0048143) |
| 1.7 | 5.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.7 | 5.2 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 1.7 | 13.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.7 | 11.9 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 1.6 | 6.5 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.6 | 9.6 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 1.6 | 4.8 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.6 | 4.7 | GO:0061552 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.5 | 12.4 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 1.5 | 4.6 | GO:1990737 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.5 | 6.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 1.5 | 4.5 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 1.5 | 4.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.5 | 5.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.4 | 11.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.4 | 15.8 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 1.4 | 18.4 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 1.4 | 4.2 | GO:0010070 | zygote asymmetric cell division(GO:0010070) |
| 1.4 | 8.5 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 1.4 | 4.2 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.4 | 4.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 1.4 | 31.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 1.4 | 5.5 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 1.4 | 5.5 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.4 | 5.5 | GO:0072697 | protein localization to cell cortex(GO:0072697) negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) negative regulation of p38MAPK cascade(GO:1903753) |
| 1.4 | 17.8 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.3 | 6.6 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 1.3 | 11.9 | GO:0099638 | endosome to plasma membrane protein transport(GO:0099638) |
| 1.3 | 16.9 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 1.3 | 3.9 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 1.3 | 3.9 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 1.3 | 3.8 | GO:0017143 | insecticide metabolic process(GO:0017143) |
| 1.3 | 11.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.3 | 3.8 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 1.3 | 3.8 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 1.2 | 23.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 1.2 | 13.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.2 | 2.5 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 1.2 | 3.7 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 1.2 | 10.9 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 1.2 | 4.8 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 1.2 | 10.7 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 1.2 | 5.9 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.2 | 3.5 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 1.1 | 3.4 | GO:0032912 | endodermal cell fate determination(GO:0007493) negative regulation of transforming growth factor beta2 production(GO:0032912) positive regulation of cardioblast differentiation(GO:0051891) cardiac vascular smooth muscle cell differentiation(GO:0060947) |
| 1.1 | 6.8 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 1.1 | 4.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 1.1 | 2.2 | GO:0050893 | sensory processing(GO:0050893) |
| 1.1 | 3.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 1.0 | 5.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 1.0 | 3.1 | GO:0006408 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 1.0 | 5.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.0 | 5.1 | GO:0061364 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 1.0 | 5.1 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.0 | 12.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.0 | 6.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.0 | 4.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.0 | 4.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.9 | 3.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.9 | 14.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.9 | 15.8 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.9 | 12.0 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.9 | 7.4 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.9 | 6.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.9 | 8.9 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.9 | 9.6 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.9 | 11.3 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.9 | 3.5 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 0.9 | 2.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.9 | 2.6 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.9 | 5.1 | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.8 | 2.5 | GO:0046223 | toxin catabolic process(GO:0009407) mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) secondary metabolite catabolic process(GO:0090487) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.8 | 2.4 | GO:0014736 | negative regulation of muscle atrophy(GO:0014736) |
| 0.8 | 4.8 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.8 | 2.4 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.8 | 5.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.8 | 4.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.8 | 2.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.8 | 4.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.8 | 5.4 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.8 | 6.9 | GO:0042473 | outer ear morphogenesis(GO:0042473) |
| 0.8 | 9.9 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.8 | 5.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.8 | 3.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.7 | 10.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.7 | 3.7 | GO:0001554 | luteolysis(GO:0001554) |
| 0.7 | 1.5 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.7 | 2.9 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.7 | 6.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.7 | 5.1 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.7 | 2.9 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.7 | 4.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.7 | 5.6 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.7 | 7.7 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.7 | 2.1 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.7 | 2.8 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.7 | 2.8 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.7 | 8.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.7 | 8.8 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.7 | 2.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.6 | 1.3 | GO:0071504 | cellular response to heparin(GO:0071504) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.6 | 9.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.6 | 5.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.6 | 3.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.6 | 5.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.6 | 1.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.6 | 1.7 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.5 | 1.6 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.5 | 6.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.5 | 3.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.5 | 1.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.5 | 2.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 1.1 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.5 | 1.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 6.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.5 | 2.6 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.5 | 2.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.5 | 3.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.5 | 4.6 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.5 | 3.0 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.5 | 6.0 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.5 | 9.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.5 | 5.8 | GO:0007614 | short-term memory(GO:0007614) |
| 0.5 | 1.9 | GO:0014028 | notochord formation(GO:0014028) |
| 0.5 | 2.9 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.5 | 6.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.5 | 3.7 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.5 | 1.9 | GO:0061107 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.5 | 8.8 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.5 | 5.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.5 | 1.8 | GO:2000553 | positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.5 | 1.4 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.5 | 6.4 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.4 | 4.0 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.4 | 4.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 7.5 | GO:0030825 | positive regulation of cGMP metabolic process(GO:0030825) |
| 0.4 | 2.6 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.4 | 3.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.4 | 5.8 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.4 | 2.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.4 | 5.4 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.4 | 8.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.4 | 9.0 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 2.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 1.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 12.7 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.4 | 3.5 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.4 | 1.2 | GO:0042891 | antibiotic transport(GO:0042891) intestinal epithelial structure maintenance(GO:0060729) |
| 0.4 | 5.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 3.5 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.4 | 4.2 | GO:0009415 | response to water(GO:0009415) |
| 0.4 | 5.3 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.4 | 11.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 3.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.4 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.4 | 7.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.4 | 25.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.4 | 1.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.4 | 1.1 | GO:0070640 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D3 metabolic process(GO:0070640) |
| 0.3 | 18.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.3 | 3.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.3 | 1.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.3 | 4.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.3 | 11.5 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) |
| 0.3 | 0.7 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.3 | 3.2 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.3 | 2.6 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 11.8 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.3 | 4.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 26.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.3 | 4.9 | GO:0060384 | innervation(GO:0060384) |
| 0.3 | 1.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.3 | 1.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.3 | 3.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 6.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 33.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 5.1 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.3 | 4.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.3 | 1.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 2.4 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.3 | 1.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 7.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 0.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 1.2 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 4.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 1.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 2.5 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.2 | 1.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 0.9 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) actin filament bundle distribution(GO:0070650) |
| 0.2 | 2.2 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 3.7 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 4.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 1.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 3.4 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 3.8 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.2 | 4.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 9.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.2 | 1.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 15.7 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.2 | 6.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 0.9 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.2 | 2.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.2 | 1.5 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.2 | 1.0 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.2 | 2.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 1.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 2.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 0.9 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 2.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.3 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 3.3 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 4.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 0.9 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.1 | 1.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.9 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 0.5 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 | 0.4 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) |
| 0.1 | 2.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 7.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.1 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 2.1 | GO:1903859 | regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 3.5 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 4.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 2.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 4.2 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 6.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.9 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 3.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.4 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.1 | 0.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 2.2 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 5.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.4 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.1 | 0.3 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 8.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.8 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 11.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 6.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.7 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 4.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.2 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.1 | 1.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.6 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 1.8 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 5.1 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 2.4 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.2 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 4.8 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.1 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 3.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.3 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 6.4 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 3.6 | GO:0035113 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 4.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 4.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 1.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.5 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 3.5 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 1.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.2 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 1.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.0 | 0.6 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.6 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.1 | GO:0007596 | blood coagulation(GO:0007596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 4.3 | 17.4 | GO:0044307 | dendritic branch(GO:0044307) |
| 4.3 | 12.9 | GO:0060187 | cell pole(GO:0060187) |
| 3.0 | 9.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 3.0 | 9.0 | GO:0035577 | azurophil granule membrane(GO:0035577) tertiary granule membrane(GO:0070821) |
| 2.3 | 9.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 2.3 | 16.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 2.2 | 6.5 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 2.1 | 10.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 2.1 | 6.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 1.8 | 28.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.8 | 19.8 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.7 | 6.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.5 | 1.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.5 | 5.8 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 1.4 | 8.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.4 | 5.5 | GO:0044393 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 1.3 | 3.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.3 | 15.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 1.3 | 5.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.2 | 3.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 1.2 | 26.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.2 | 9.4 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.1 | 6.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.1 | 6.8 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 1.1 | 8.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.1 | 8.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.1 | 5.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.1 | 3.2 | GO:0032173 | septin collar(GO:0032173) |
| 1.0 | 3.8 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.0 | 3.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.0 | 6.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.9 | 7.4 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.9 | 11.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.8 | 3.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.8 | 4.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.8 | 10.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.8 | 84.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.8 | 3.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.8 | 15.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.7 | 3.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.7 | 3.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.7 | 4.0 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.7 | 7.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.7 | 14.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.6 | 6.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.6 | 15.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.6 | 3.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.6 | 1.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 1.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.5 | 1.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 2.1 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.5 | 5.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.5 | 3.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.5 | 2.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.5 | 9.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.5 | 2.9 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.5 | 3.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.4 | 3.8 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.4 | 8.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.4 | 6.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 3.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 14.4 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 5.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 7.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.4 | 10.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 9.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 1.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.3 | 14.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 11.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 3.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.3 | 2.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 2.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 3.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 5.5 | GO:0098793 | presynapse(GO:0098793) |
| 0.3 | 5.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 5.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.3 | 2.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.3 | 3.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 43.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 6.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 5.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 3.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 2.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 40.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 40.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 9.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 9.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.2 | 13.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 5.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 2.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.0 | GO:1903767 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
| 0.2 | 4.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 1.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 5.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 20.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 11.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 2.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 5.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 10.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 16.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 11.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 8.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 22.7 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 6.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 5.8 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 3.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 9.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.2 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.1 | 9.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 2.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 107.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 4.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 20.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.6 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 32.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 5.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 13.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 8.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 11.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 35.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 2.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 7.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 5.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 16.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 9.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 11.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 24.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 98.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 12.9 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 16.8 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0071944 | cell periphery(GO:0071944) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 30.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 5.6 | 22.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 5.5 | 43.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 4.4 | 22.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 4.0 | 16.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 4.0 | 12.0 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 3.9 | 23.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 3.8 | 18.8 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 3.3 | 9.8 | GO:0038100 | nodal binding(GO:0038100) |
| 3.2 | 12.9 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 3.1 | 12.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.9 | 14.6 | GO:1904315 | transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.9 | 8.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 2.9 | 8.6 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 2.8 | 8.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 2.8 | 8.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 2.2 | 8.9 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 2.2 | 11.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 2.1 | 6.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 2.0 | 16.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.9 | 5.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.9 | 5.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.9 | 5.7 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 1.8 | 11.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 1.7 | 10.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.7 | 6.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 1.7 | 10.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.6 | 6.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 1.6 | 9.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.6 | 14.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.6 | 15.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.6 | 14.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.5 | 4.6 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 1.5 | 4.5 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 1.5 | 5.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.4 | 9.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 1.4 | 13.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.3 | 6.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.3 | 9.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.3 | 5.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.2 | 3.7 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 1.2 | 31.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 1.2 | 5.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.1 | 6.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 1.1 | 5.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.1 | 37.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.1 | 5.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 1.1 | 4.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.1 | 14.2 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 1.1 | 3.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 1.1 | 9.5 | GO:0070061 | fructose binding(GO:0070061) |
| 1.0 | 5.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 1.0 | 28.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.0 | 13.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.0 | 5.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.0 | 5.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 1.0 | 11.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.9 | 8.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.9 | 4.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.9 | 4.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.9 | 9.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.9 | 5.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.9 | 10.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.9 | 2.6 | GO:0052743 | inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.9 | 5.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.8 | 15.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.8 | 1.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.8 | 5.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.8 | 3.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.8 | 3.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.8 | 2.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.8 | 4.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.8 | 3.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.8 | 14.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.8 | 6.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.7 | 6.0 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.7 | 10.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.7 | 3.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.7 | 2.8 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.7 | 2.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.7 | 3.5 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.7 | 4.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.7 | 5.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.7 | 2.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.7 | 15.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.7 | 28.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.7 | 28.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.6 | 4.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 5.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 9.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.6 | 4.4 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.6 | 7.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.6 | 5.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.6 | 2.4 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.6 | 15.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.6 | 3.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.6 | 4.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.6 | 1.7 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.6 | 8.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.6 | 1.7 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.5 | 2.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.5 | 6.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.5 | 4.4 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.5 | 6.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.5 | 3.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 5.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 16.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 5.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 4.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.4 | 1.6 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.4 | 2.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.4 | 1.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.4 | 14.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 9.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.4 | 4.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 2.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.4 | 3.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 3.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.4 | 1.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.4 | 1.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 4.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 7.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 5.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 12.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.3 | 5.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 2.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 2.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 16.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 1.0 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.3 | 1.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.3 | 6.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 3.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.3 | 7.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 12.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 3.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 3.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 4.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 0.9 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.3 | 3.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 3.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 7.5 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.3 | 18.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.3 | 10.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 1.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.3 | 8.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 4.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 11.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 4.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 22.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 3.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.2 | 2.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 2.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 23.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 19.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 2.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 7.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 7.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 4.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 1.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 5.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 5.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 3.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 0.9 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 3.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 7.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 2.0 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 1.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 1.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 3.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 5.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 4.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 3.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 6.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 12.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 2.7 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 5.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 2.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 9.2 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.1 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 3.5 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 6.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 2.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 11.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 0.4 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 13.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 3.0 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 1.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.7 | GO:0052745 | inositol phosphate phosphatase activity(GO:0052745) |
| 0.1 | 1.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 2.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 2.2 | GO:0004601 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.1 | 0.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.5 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.1 | 1.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 2.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 4.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 4.0 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 20.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.3 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.1 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 2.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 2.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 1.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.3 | GO:0035254 | glutamate receptor binding(GO:0035254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 1.3 | 27.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 1.0 | 18.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 1.0 | 20.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.8 | 20.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.7 | 17.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.7 | 20.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.7 | 9.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.5 | 9.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.5 | 6.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 8.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 4.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 16.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.4 | 7.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 9.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 10.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.4 | 7.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.4 | 19.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 5.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 19.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 10.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.3 | 6.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 9.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 78.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 5.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 4.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 14.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 6.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 8.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 31.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 2.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 5.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 34.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 1.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 3.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 3.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 5.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 8.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 4.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 4.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 40.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.6 | 21.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.4 | 29.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 1.2 | 27.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 1.1 | 18.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 1.0 | 31.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 1.0 | 15.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.9 | 26.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.9 | 13.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.9 | 12.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.8 | 13.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.8 | 4.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.8 | 9.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.7 | 6.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.7 | 21.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.7 | 23.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.6 | 9.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.6 | 4.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.6 | 13.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 9.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 9.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.5 | 6.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.5 | 16.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.5 | 4.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.5 | 7.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.5 | 3.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.5 | 3.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.5 | 38.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.5 | 5.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.5 | 19.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 4.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 8.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 5.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 4.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 4.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 1.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.4 | 34.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.4 | 15.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 10.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 5.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 4.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 3.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.3 | 3.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.3 | 10.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 5.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 10.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 4.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 5.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 3.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 8.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 3.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 6.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 4.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 5.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 3.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 2.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 5.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 3.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.2 | 10.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 7.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 2.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 6.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 3.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 4.6 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.2 | 3.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 4.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 4.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 24.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 2.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 2.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 6.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 9.9 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.2 | 13.9 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.2 | 2.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 22.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 8.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 3.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 5.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 5.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 3.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.0 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 2.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |