Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
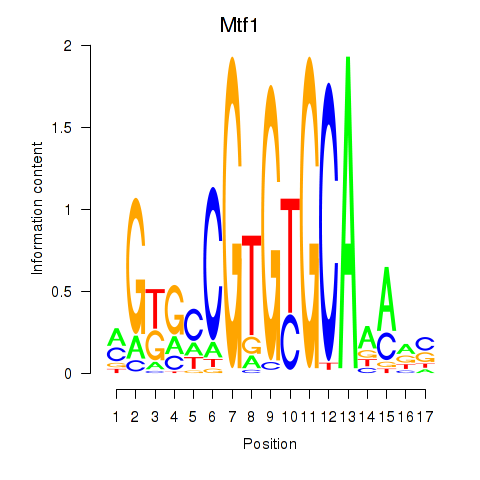
Results for Mtf1
Z-value: 0.66
Transcription factors associated with Mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mtf1
|
ENSRNOG00000025724 | metal-regulatory transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mtf1 | rn6_v1_chr5_+_142797366_142797366 | 0.13 | 1.7e-02 | Click! |
Activity profile of Mtf1 motif
Sorted Z-values of Mtf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11302938 | 46.34 |
ENSRNOT00000038212
|
AC128848.1
|
|
| chr19_-_11308740 | 46.16 |
ENSRNOT00000067391
|
Mt2A
|
metallothionein 2A |
| chr17_+_43423111 | 18.37 |
ENSRNOT00000022630
|
Hist1h2ba
|
histone cluster 1 H2B family member a |
| chr9_+_93030714 | 17.71 |
ENSRNOT00000023581
|
Spata3
|
spermatogenesis associated 3 |
| chrX_+_74497262 | 14.35 |
ENSRNOT00000003899
|
Zcchc13
|
zinc finger CCHC-type containing 13 |
| chr7_-_117679219 | 12.99 |
ENSRNOT00000071522
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr8_+_49342067 | 12.65 |
ENSRNOT00000021693
|
Mpzl2
|
myelin protein zero-like 2 |
| chr7_-_117680004 | 11.82 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr1_-_81377976 | 11.74 |
ENSRNOT00000075189
|
Srrm5
|
serine/arginine repetitive matrix 5 |
| chr1_+_189549960 | 10.82 |
ENSRNOT00000019654
|
Exnef
|
exonuclease NEF-sp |
| chr13_+_110677810 | 10.73 |
ENSRNOT00000006340
|
Slc30a1
|
solute carrier family 30 member 1 |
| chr1_+_217018916 | 10.67 |
ENSRNOT00000028195
ENSRNOT00000078979 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr20_-_5806097 | 10.50 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr1_-_213921208 | 10.37 |
ENSRNOT00000044393
|
Ano9
|
anoctamin 9 |
| chr1_+_72882806 | 10.33 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr13_+_75059927 | 10.12 |
ENSRNOT00000080801
|
LOC680254
|
hypothetical protein LOC680254 |
| chr9_-_119871382 | 9.25 |
ENSRNOT00000018644
|
Ndc80
|
NDC80 kinetochore complex component |
| chr16_+_47368768 | 8.91 |
ENSRNOT00000017966
|
Wwc2
|
WW and C2 domain containing 2 |
| chr16_-_21348391 | 8.88 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr6_+_127400585 | 8.70 |
ENSRNOT00000087429
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr6_-_47811853 | 8.65 |
ENSRNOT00000010942
|
Allc
|
allantoicase |
| chrX_+_29157470 | 8.35 |
ENSRNOT00000081986
|
LOC100910245
|
ribose-phosphate pyrophosphokinase 2-like |
| chr19_+_25983169 | 8.24 |
ENSRNOT00000004404
|
Syce2
|
synaptonemal complex central element protein 2 |
| chr5_+_152559577 | 8.22 |
ENSRNOT00000082245
|
Slc30a2
|
solute carrier family 30 member 2 |
| chr13_-_84217332 | 8.14 |
ENSRNOT00000047488
ENSRNOT00000087096 |
Pou2f1
|
POU class 2 homeobox 1 |
| chr1_-_219853329 | 8.08 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr19_+_10142496 | 7.97 |
ENSRNOT00000088645
ENSRNOT00000060351 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr1_-_81127059 | 6.46 |
ENSRNOT00000026252
|
Zfp94
|
zinc finger protein 94 |
| chr4_+_78458625 | 6.42 |
ENSRNOT00000049891
|
Tmem176a
|
transmembrane protein 176A |
| chr4_+_170149029 | 6.36 |
ENSRNOT00000073287
|
LOC103690002
|
histone H2A.J |
| chrX_+_28435507 | 6.31 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr4_-_66899914 | 6.30 |
ENSRNOT00000011481
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr5_-_147743723 | 6.26 |
ENSRNOT00000012854
|
Hdac1
|
histone deacetylase 1 |
| chr2_-_45077219 | 6.23 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr6_-_26875376 | 6.22 |
ENSRNOT00000011838
|
Tmem214
|
transmembrane protein 214 |
| chr3_-_171166454 | 6.22 |
ENSRNOT00000034131
|
Ctcfl
|
CCCTC-binding factor like |
| chr2_-_27364906 | 6.19 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr20_-_5805627 | 5.99 |
ENSRNOT00000085996
|
Clps
|
colipase |
| chr9_-_84894599 | 5.94 |
ENSRNOT00000018423
|
Hdac1l
|
histone deacetylase 1-like |
| chr3_+_152143811 | 5.94 |
ENSRNOT00000026578
|
LOC100911109
|
sperm-associated antigen 4 protein-like |
| chr7_-_141307233 | 5.76 |
ENSRNOT00000071885
|
Racgap1
|
Rac GTPase-activating protein 1 |
| chr1_-_220096319 | 5.68 |
ENSRNOT00000091787
ENSRNOT00000073983 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr8_-_71784551 | 5.66 |
ENSRNOT00000022960
|
Snx1
|
sorting nexin 1 |
| chr3_+_151609602 | 5.62 |
ENSRNOT00000065052
|
Spag4
|
sperm associated antigen 4 |
| chr11_-_90406797 | 5.01 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr10_-_78219793 | 4.95 |
ENSRNOT00000003369
|
Tom1l1
|
target of myb1 like 1 membrane trafficking protein |
| chr11_+_67757928 | 4.85 |
ENSRNOT00000039215
|
Dtx3l
|
deltex E3 ubiquitin ligase 3L |
| chr4_+_7355574 | 4.84 |
ENSRNOT00000013800
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr1_-_89042176 | 4.72 |
ENSRNOT00000080842
|
Kmt2b
|
lysine methyltransferase 2B |
| chr4_-_115453659 | 4.59 |
ENSRNOT00000065847
|
Tex261
|
testis expressed 261 |
| chr8_+_60117729 | 4.53 |
ENSRNOT00000021074
|
Isl2
|
ISL LIM homeobox 2 |
| chr19_+_55300395 | 4.52 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr16_-_40555576 | 4.46 |
ENSRNOT00000015529
|
Vegfc
|
vascular endothelial growth factor C |
| chr7_-_116963281 | 4.21 |
ENSRNOT00000082058
ENSRNOT00000012306 |
Tsta3
|
tissue specific transplantation antigen P35B |
| chr2_+_27365148 | 4.12 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr2_+_198359754 | 3.82 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr5_-_74190991 | 3.74 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr12_+_7454884 | 3.47 |
ENSRNOT00000077328
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr20_-_35450513 | 3.45 |
ENSRNOT00000087342
|
Man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr19_-_55300403 | 3.09 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr11_+_81757983 | 3.01 |
ENSRNOT00000088503
|
Tbccd1
|
TBCC domain containing 1 |
| chr1_-_189549891 | 2.91 |
ENSRNOT00000019747
|
Eri2
|
ERI1 exoribonuclease family member 2 |
| chr12_-_2545740 | 2.90 |
ENSRNOT00000075527
|
Tgfbr3l
|
transforming growth factor beta receptor 3 like |
| chr16_+_7035068 | 2.66 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chr8_-_114892910 | 2.09 |
ENSRNOT00000074934
|
Ppm1m
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr5_-_58484900 | 2.09 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr3_+_175548174 | 2.07 |
ENSRNOT00000091941
|
Adrm1
|
adhesion regulating molecule 1 |
| chr12_-_11265865 | 1.99 |
ENSRNOT00000001315
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr12_+_52359310 | 1.95 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr5_+_164950917 | 1.73 |
ENSRNOT00000083678
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr9_+_16543688 | 1.71 |
ENSRNOT00000021868
|
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr16_+_71629525 | 1.67 |
ENSRNOT00000035347
ENSRNOT00000088462 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr5_+_164951789 | 1.52 |
ENSRNOT00000055658
ENSRNOT00000012193 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr16_+_20109200 | 1.48 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr11_-_81757813 | 1.48 |
ENSRNOT00000002462
|
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr13_+_97702097 | 1.27 |
ENSRNOT00000057787
|
LOC103689999
|
saccharopine dehydrogenase-like oxidoreductase |
| chr8_-_62948720 | 1.21 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr3_-_9448167 | 1.16 |
ENSRNOT00000012617
|
Abl1
|
ABL proto-oncogene 1, non-receptor tyrosine kinase |
| chr3_-_13978224 | 1.11 |
ENSRNOT00000025528
|
Phf19
|
PHD finger protein 19 |
| chr10_-_18574909 | 1.09 |
ENSRNOT00000083090
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chrX_-_16526028 | 0.96 |
ENSRNOT00000093647
|
Dgkk
|
diacylglycerol kinase kappa |
| chr2_-_225389120 | 0.89 |
ENSRNOT00000016739
|
Abcd3
|
ATP binding cassette subfamily D member 3 |
| chr20_-_422464 | 0.79 |
ENSRNOT00000051646
|
Olr1673
|
olfactory receptor 1673 |
| chr1_+_29432152 | 0.76 |
ENSRNOT00000019436
ENSRNOT00000083276 |
Trmt11
|
tRNA methyltransferase 11 homolog |
| chr3_-_9447813 | 0.69 |
ENSRNOT00000043661
|
Abl1
|
ABL proto-oncogene 1, non-receptor tyrosine kinase |
| chr4_+_144916789 | 0.55 |
ENSRNOT00000083660
|
AABR07061801.2
|
|
| chr4_-_123494742 | 0.50 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr20_-_529024 | 0.44 |
ENSRNOT00000075114
|
Olr1675
|
olfactory receptor 1675 |
| chr5_-_106758032 | 0.41 |
ENSRNOT00000033651
|
Hacd4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr19_-_52282877 | 0.36 |
ENSRNOT00000021271
|
Kcng4
|
potassium voltage-gated channel modifier subfamily G member 4 |
| chr16_+_7035241 | 0.24 |
ENSRNOT00000084977
|
Nek4
|
NIMA-related kinase 4 |
| chr4_+_175431904 | 0.03 |
ENSRNOT00000032843
|
Pde3a
|
phosphodiesterase 3A |
| chr20_-_11737050 | 0.03 |
ENSRNOT00000001637
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chrX_+_10964067 | 0.03 |
ENSRNOT00000093181
ENSRNOT00000066480 |
Med14
|
mediator complex subunit 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mtf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 24.8 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 6.6 | 46.2 | GO:0048143 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) astrocyte activation(GO:0048143) |
| 3.6 | 10.7 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 3.6 | 10.7 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 3.5 | 10.4 | GO:2001226 | negative regulation of inorganic anion transmembrane transport(GO:1903796) negative regulation of chloride transport(GO:2001226) |
| 3.1 | 18.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 2.9 | 8.6 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 2.0 | 8.1 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.9 | 5.8 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 1.7 | 5.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.6 | 4.8 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 1.6 | 6.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 1.6 | 6.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.6 | 6.3 | GO:0061197 | fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 1.5 | 4.5 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 1.5 | 4.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 1.5 | 4.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 1.5 | 8.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.4 | 4.2 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 1.2 | 9.2 | GO:0090267 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.0 | 2.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.9 | 8.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.9 | 6.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.8 | 4.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.8 | 3.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.8 | 3.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.7 | 8.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.7 | 5.6 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.6 | 10.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.6 | 1.8 | GO:0002332 | transitional stage B cell differentiation(GO:0002332) transitional one stage B cell differentiation(GO:0002333) positive regulation of actin filament binding(GO:1904531) positive regulation of actin binding(GO:1904618) |
| 0.5 | 1.5 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.4 | 2.9 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.4 | 3.7 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.4 | 5.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.4 | 8.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 10.9 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.3 | 8.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.3 | 8.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 3.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 5.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.2 | 2.9 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.2 | 16.5 | GO:0032094 | response to food(GO:0032094) |
| 0.2 | 5.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 4.8 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.2 | 5.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 3.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 11.7 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.9 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 1.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 8.4 | GO:0009156 | ribonucleoside monophosphate biosynthetic process(GO:0009156) |
| 0.1 | 6.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.5 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 1.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 2.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.1 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 10.8 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 3.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 2.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.8 | 9.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) Ndc80 complex(GO:0031262) |
| 1.4 | 5.8 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.3 | 6.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.7 | 8.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.7 | 3.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.6 | 8.2 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 24.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.4 | 10.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 6.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 25.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 2.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 3.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 5.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 2.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 8.7 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) |
| 0.1 | 10.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 5.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 4.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 6.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 5.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 8.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 8.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 47.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 5.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 6.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 3.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 10.8 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 46.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 2.6 | 10.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.1 | 6.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.9 | 5.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.8 | 14.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.7 | 43.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 1.1 | 5.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.8 | 8.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.7 | 4.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.7 | 4.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.7 | 8.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 5.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.5 | 10.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 4.5 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.4 | 10.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.4 | 4.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 2.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 8.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.3 | 3.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 2.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 6.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 3.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 0.8 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 3.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 5.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 6.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 4.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 0.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 15.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 10.8 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 8.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 8.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 5.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 10.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 26.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 5.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.2 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 5.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 6.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 18.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 46.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.5 | 6.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.4 | 4.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 16.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 8.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 8.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 8.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 5.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 5.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 43.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.8 | 45.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.7 | 18.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.5 | 10.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 16.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 4.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 8.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.3 | 10.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 6.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 5.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 4.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 5.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 9.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 9.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 3.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 6.3 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |