Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
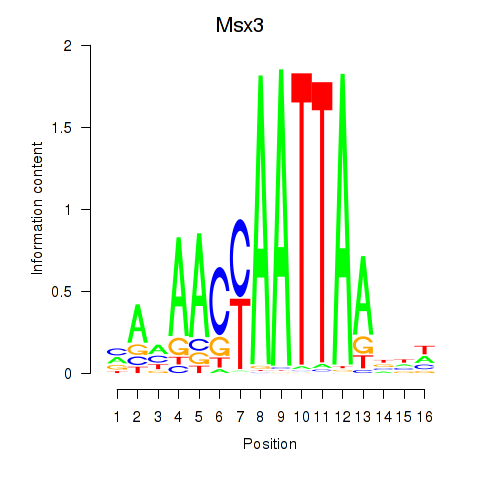
Results for Msx3
Z-value: 0.42
Transcription factors associated with Msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx3
|
ENSRNOG00000046776 | msh homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx3 | rn6_v1_chr1_-_212517151_212517151 | -0.25 | 7.4e-06 | Click! |
Activity profile of Msx3 motif
Sorted Z-values of Msx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_69404323 | 12.03 |
ENSRNOT00000051342
ENSRNOT00000066282 |
Akr1c2
|
aldo-keto reductase family 1, member C2 |
| chr5_+_28485619 | 11.73 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr8_+_48718329 | 10.93 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr1_+_185863043 | 10.14 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr10_+_84135116 | 8.83 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr9_-_18371856 | 8.69 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr6_-_42002819 | 8.51 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr8_-_119012671 | 8.38 |
ENSRNOT00000028435
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr16_-_24951612 | 8.02 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr3_-_165700489 | 7.74 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr13_-_111972603 | 7.62 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr3_-_165537940 | 7.42 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr18_-_13183263 | 7.02 |
ENSRNOT00000050933
|
Ccdc178
|
coiled-coil domain containing 178 |
| chrX_-_98095663 | 6.77 |
ENSRNOT00000004491
|
Rbm31y
|
RNA binding motif 31, Y-linked |
| chr8_+_128027958 | 6.40 |
ENSRNOT00000045049
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr18_+_45023932 | 6.07 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr1_+_101603222 | 5.50 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr1_-_14117021 | 5.24 |
ENSRNOT00000004344
|
RGD1560303
|
similar to hypothetical protein 4933423E17 |
| chr4_-_178168690 | 5.13 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr4_+_147832136 | 4.99 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr6_-_86223052 | 4.84 |
ENSRNOT00000046828
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr7_-_143167772 | 4.44 |
ENSRNOT00000011374
|
Krt85
|
keratin 85 |
| chr9_+_60900213 | 4.38 |
ENSRNOT00000017859
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr20_+_32717564 | 4.22 |
ENSRNOT00000030642
|
Rfx6
|
regulatory factor X, 6 |
| chr18_+_12056113 | 3.80 |
ENSRNOT00000038450
|
Dsg4
|
desmoglein 4 |
| chr14_+_23405717 | 3.69 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr4_-_176606382 | 3.49 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr7_+_143122269 | 3.43 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr16_+_2634603 | 3.07 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr3_+_112346627 | 2.93 |
ENSRNOT00000074392
|
Snap23
|
synaptosomal-associated protein 23 |
| chr2_+_220432037 | 2.89 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr15_+_39779648 | 2.86 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chrX_+_110789269 | 2.85 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr20_+_44680449 | 2.82 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr6_+_8886591 | 2.69 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr12_-_45801842 | 2.36 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr12_-_22138382 | 2.33 |
ENSRNOT00000001899
|
Lrch4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr16_-_75241303 | 2.31 |
ENSRNOT00000058056
|
Defb2
|
defensin beta 2 |
| chr2_-_158156444 | 2.30 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_248398917 | 2.30 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr1_+_217459215 | 2.27 |
ENSRNOT00000092386
ENSRNOT00000092357 |
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr10_-_87286387 | 2.24 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chrM_+_9870 | 2.15 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr3_+_161160920 | 2.15 |
ENSRNOT00000019849
|
Spint4
|
serine protease inhibitor, Kunitz type 4 |
| chr7_-_73130740 | 2.13 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr7_-_15706893 | 2.11 |
ENSRNOT00000042742
|
Olr1092
|
olfactory receptor 1092 |
| chr5_-_115387377 | 2.09 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr5_+_6373583 | 2.09 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr7_-_143967484 | 1.72 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr1_-_172076138 | 1.71 |
ENSRNOT00000051241
|
Olr239
|
olfactory receptor 239 |
| chrM_+_9451 | 1.70 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr3_-_10196626 | 1.66 |
ENSRNOT00000012234
|
Prdm12
|
PR/SET domain 12 |
| chr6_-_36876805 | 1.60 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr10_-_87195075 | 1.58 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chr3_-_77800137 | 1.52 |
ENSRNOT00000071319
|
AABR07052831.1
|
|
| chr6_-_50618694 | 1.51 |
ENSRNOT00000008980
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr15_-_36798814 | 1.49 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr10_-_88000423 | 1.40 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr1_-_8885967 | 1.35 |
ENSRNOT00000016102
|
Gje1
|
gap junction protein, epsilon 1 |
| chr2_-_192812008 | 1.29 |
ENSRNOT00000080554
|
Lce6a
|
late cornified envelope 6A |
| chr9_-_24467892 | 1.29 |
ENSRNOT00000060803
|
Defb18
|
defensin beta 18 |
| chr6_-_111417813 | 1.10 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr5_-_56567320 | 1.08 |
ENSRNOT00000066081
|
Topors
|
TOP1 binding arginine/serine rich protein |
| chr8_+_115546893 | 1.02 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr13_-_101667147 | 0.90 |
ENSRNOT00000080475
|
AABR07021987.1
|
|
| chr10_+_82032656 | 0.86 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chrX_-_70835089 | 0.76 |
ENSRNOT00000076351
|
Tex11
|
testis expressed 11 |
| chr1_-_167971151 | 0.75 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr8_+_79638696 | 0.74 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr2_+_195996521 | 0.71 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr11_-_28979169 | 0.65 |
ENSRNOT00000061601
|
AABR07033580.1
|
|
| chr12_+_18679789 | 0.64 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr16_-_16762240 | 0.61 |
ENSRNOT00000080746
|
AABR07024790.1
|
|
| chr16_-_75028977 | 0.60 |
ENSRNOT00000058071
|
Defb13
|
defensin beta 13 |
| chr1_-_168930776 | 0.59 |
ENSRNOT00000021468
|
LOC103694854
|
olfactory receptor 52Z1-like |
| chr18_-_43945273 | 0.50 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chrY_-_1305026 | 0.49 |
ENSRNOT00000092901
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr1_+_229889771 | 0.44 |
ENSRNOT00000085941
|
Olr348
|
olfactory receptor 348 |
| chr1_-_149529350 | 0.35 |
ENSRNOT00000052226
|
Vom2r43
|
vomeronasal 2 receptor, 43 |
| chr11_-_71136673 | 0.34 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chrX_-_120521871 | 0.33 |
ENSRNOT00000080863
|
LOC100362376
|
LRRGT00025-like |
| chr1_-_101095594 | 0.32 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr10_+_53740841 | 0.29 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr8_-_39551700 | 0.25 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr4_-_66955732 | 0.24 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr3_-_30930141 | 0.23 |
ENSRNOT00000037001
|
AABR07051982.1
|
|
| chr4_+_88694583 | 0.18 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr8_+_43722228 | 0.16 |
ENSRNOT00000084016
|
Olr1325
|
olfactory receptor 1325 |
| chr1_-_57518458 | 0.01 |
ENSRNOT00000002040
|
Pdcd2
|
programmed cell death 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 2.5 | 15.3 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 2.5 | 7.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.6 | 10.9 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.9 | 2.7 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.6 | 5.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.6 | 8.4 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.6 | 11.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.6 | 1.7 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.5 | 3.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 1.0 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.5 | 1.5 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.4 | 5.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.4 | 4.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.4 | 2.9 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 8.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.4 | 2.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.4 | 1.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.4 | 7.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 1.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.3 | 3.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 8.8 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.3 | 3.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.3 | 5.2 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.2 | 2.3 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 2.3 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.2 | 0.6 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.2 | 0.8 | GO:0006311 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 0.1 | 2.8 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 1.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 2.9 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 1.7 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 1.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 1.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 2.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 1.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 4.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 6.4 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.6 | 2.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 5.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 1.5 | GO:0043159 | acrosomal matrix(GO:0043159) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 5.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 4.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 5.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 2.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 3.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 2.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 8.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 8.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 10.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 3.8 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 7.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 6.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 7.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 3.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 2.5 | 7.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.6 | 10.9 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 1.0 | 11.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.9 | 6.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.7 | 2.9 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.6 | 5.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 3.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.6 | 2.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.6 | 5.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.3 | 5.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 1.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 1.5 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 2.3 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.1 | 3.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 8.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.7 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 1.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 6.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 13.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 14.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 8.7 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 12.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 5.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 7.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 7.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 10.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 2.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 8.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.9 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 2.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |