Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Msx2_Hoxd4
Z-value: 0.35
Transcription factors associated with Msx2_Hoxd4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
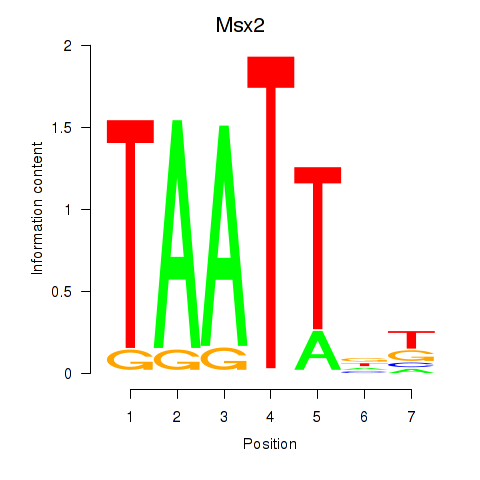
Msx2
|
ENSRNOG00000018355 | msh homeobox 2 |
|
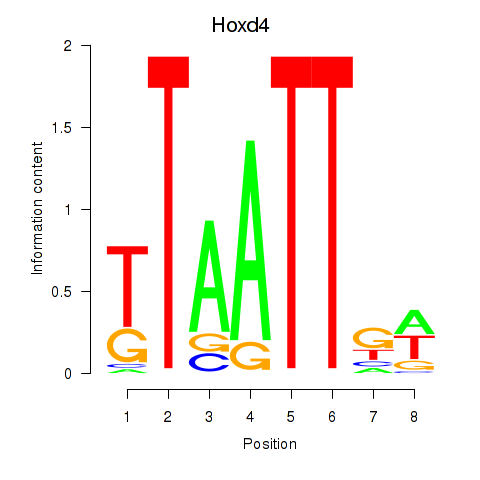
Hoxd4
|
ENSRNOG00000001578 | homeo box D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx2 | rn6_v1_chr17_+_11683862_11683862 | 0.48 | 6.5e-20 | Click! |
Activity profile of Msx2_Hoxd4 motif
Sorted Z-values of Msx2_Hoxd4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_47094352 | 11.65 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chr20_-_25826658 | 7.78 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr6_+_64789940 | 7.67 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr15_-_71779033 | 6.14 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr7_+_44009069 | 5.94 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr4_-_64330996 | 5.82 |
ENSRNOT00000016088
|
Ptn
|
pleiotrophin |
| chrX_+_84064427 | 5.51 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr11_+_61605937 | 4.85 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
| chr5_-_168734296 | 4.59 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr2_-_35104963 | 4.56 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr9_+_73418607 | 4.42 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr9_-_30844199 | 4.35 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr7_-_69982592 | 3.90 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr3_+_61613774 | 3.59 |
ENSRNOT00000002148
|
Hoxd10
|
homeo box D10 |
| chr1_+_47032113 | 3.41 |
ENSRNOT00000024412
|
Tulp4
|
tubby like protein 4 |
| chr3_-_52447622 | 3.20 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr12_+_39423596 | 3.01 |
ENSRNOT00000066952
|
Ift81
|
intraflagellar transport 81 |
| chrX_+_908044 | 2.96 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr3_-_25212049 | 2.91 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr18_+_54108493 | 2.80 |
ENSRNOT00000029239
|
RGD1312005
|
similar to DD1 |
| chr13_-_76049363 | 2.70 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr5_+_104698040 | 2.61 |
ENSRNOT00000009127
|
Adamtsl1
|
ADAMTS-like 1 |
| chr11_-_782954 | 2.59 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr2_+_127489771 | 2.49 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr20_+_25990656 | 2.39 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr6_-_76552559 | 2.38 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr2_-_210766501 | 2.38 |
ENSRNOT00000025800
|
Gstm3
|
glutathione S-transferase mu 3 |
| chr13_-_86671515 | 2.31 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr18_-_26656879 | 2.06 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr13_-_27192592 | 1.95 |
ENSRNOT00000040021
|
Serpinb3
|
serpin family B member 3 |
| chr4_+_24222500 | 1.80 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr9_-_63641400 | 1.75 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr2_-_181531978 | 1.64 |
ENSRNOT00000072029
|
Npy2r
|
neuropeptide Y receptor Y2 |
| chr10_+_65733991 | 1.62 |
ENSRNOT00000013698
|
Slc46a1
|
solute carrier family 46 member 1 |
| chr15_+_32894938 | 1.48 |
ENSRNOT00000012837
|
Abhd4
|
abhydrolase domain containing 4 |
| chr12_-_35979193 | 1.46 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr13_+_46169963 | 1.34 |
ENSRNOT00000005212
|
Thsd7b
|
thrombospondin type 1 domain containing 7B |
| chr15_+_56666012 | 1.32 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr4_+_31333970 | 1.29 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr7_+_35773928 | 1.23 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr5_-_128446725 | 1.20 |
ENSRNOT00000011272
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr8_-_43192910 | 1.02 |
ENSRNOT00000060101
|
Olr1302
|
olfactory receptor 1302 |
| chr9_+_10941613 | 1.01 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr7_-_143392777 | 1.00 |
ENSRNOT00000086504
ENSRNOT00000038105 |
Krt72
|
keratin 72 |
| chr8_+_56585396 | 0.95 |
ENSRNOT00000085316
ENSRNOT00000034924 |
Rdx
|
radixin |
| chr10_-_87232723 | 0.90 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr6_-_94834908 | 0.85 |
ENSRNOT00000006284
|
L3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr2_+_196013799 | 0.85 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr2_+_72006099 | 0.84 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr16_-_24951612 | 0.80 |
ENSRNOT00000018987
|
Tktl2
|
transketolase-like 2 |
| chr4_-_157304653 | 0.77 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr19_-_22194740 | 0.76 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr2_-_185005572 | 0.69 |
ENSRNOT00000093291
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr2_+_154604832 | 0.64 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr8_+_43705216 | 0.62 |
ENSRNOT00000090425
|
AC115384.1
|
|
| chr4_-_78879294 | 0.59 |
ENSRNOT00000084543
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chrX_-_106607352 | 0.58 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr1_+_150310319 | 0.55 |
ENSRNOT00000042081
|
Olr34
|
olfactory receptor 34 |
| chr6_+_101532518 | 0.47 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr8_+_43747548 | 0.45 |
ENSRNOT00000092067
|
Olr1327
|
olfactory receptor 1327 |
| chr19_+_25043680 | 0.44 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr19_+_43163129 | 0.44 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr13_+_90533365 | 0.43 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr1_+_279633671 | 0.35 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr13_-_32427177 | 0.35 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr2_-_33025271 | 0.35 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_-_99214251 | 0.34 |
ENSRNOT00000042682
|
Esr2
|
estrogen receptor 2 |
| chr4_-_129619142 | 0.32 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr1_+_169115981 | 0.25 |
ENSRNOT00000067478
|
Olr135
|
olfactory receptor 135 |
| chr7_+_122818975 | 0.22 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chr1_+_79474809 | 0.17 |
ENSRNOT00000044927
|
Ceacam12
|
carcinoembryonic antigen-related cell adhesion molecule 12 |
| chr1_+_168390615 | 0.13 |
ENSRNOT00000039822
|
Olr87
|
olfactory receptor 87 |
| chr17_+_45670284 | 0.13 |
ENSRNOT00000086536
|
Olr1660
|
olfactory receptor 1660 |
| chr6_-_146902339 | 0.12 |
ENSRNOT00000050893
|
Abcb5
|
ATP binding cassette subfamily B member 5 |
| chr3_-_74014795 | 0.11 |
ENSRNOT00000073426
|
Olr516
|
olfactory receptor 516 |
| chr7_-_143408276 | 0.09 |
ENSRNOT00000013122
ENSRNOT00000091540 |
Krt73
|
keratin 73 |
| chr19_+_32308236 | 0.09 |
ENSRNOT00000015508
|
Mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr4_+_8066737 | 0.07 |
ENSRNOT00000063994
|
Srpk2
|
SRSF protein kinase 2 |
| chr1_-_56683731 | 0.01 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx2_Hoxd4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | GO:1904373 | retinal rod cell differentiation(GO:0060221) response to kainic acid(GO:1904373) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.0 | 7.8 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.9 | 2.6 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.9 | 7.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.8 | 2.4 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.6 | 1.9 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.4 | 1.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.4 | 1.6 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.4 | 2.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.3 | 3.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 11.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 3.6 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 1.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 1.6 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.2 | 2.4 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 | 1.0 | GO:1902966 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 | 1.7 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 2.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 4.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.3 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) cellular response to magnetism(GO:0071259) |
| 0.1 | 3.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.2 | GO:0018076 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 7.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 2.7 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 2.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 2.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 4.4 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.6 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 4.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 7.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 10.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 3.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.0 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 5.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 3.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 15.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 1.7 | 11.6 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.5 | 5.8 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.4 | 1.3 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.4 | 1.6 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.3 | 1.6 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.3 | 2.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 4.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.5 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.1 | 3.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.3 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 5.9 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 6.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 4.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 5.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 7.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 4.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |