Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Msx1_Lhx9_Barx1_Rax_Dlx6
Z-value: 0.43
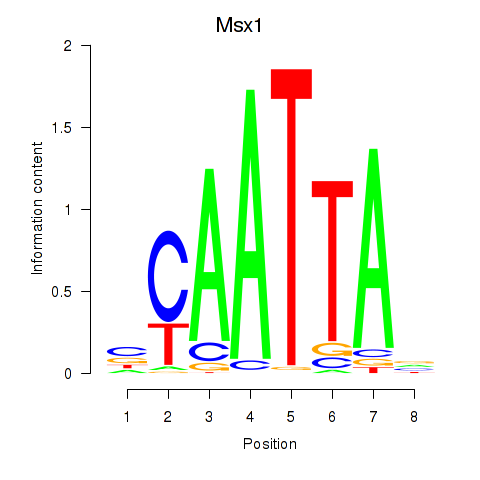
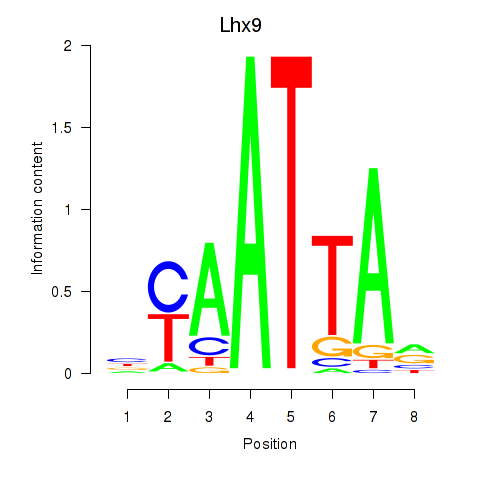
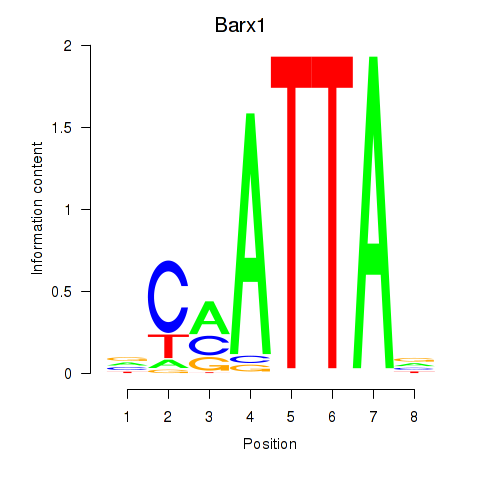
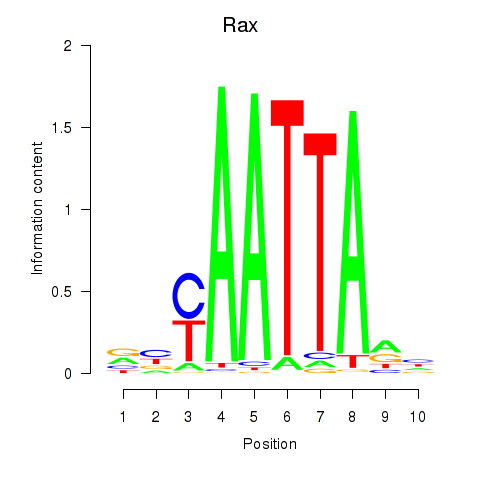
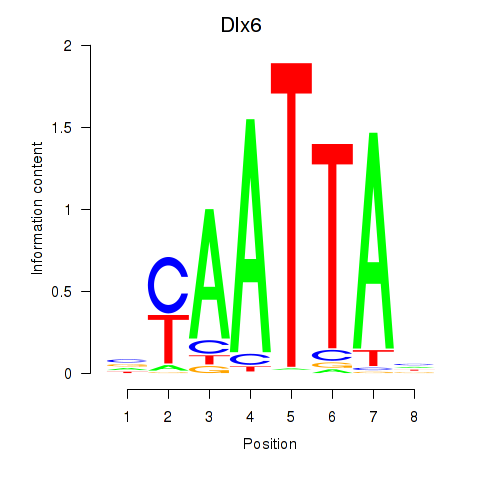
Transcription factors associated with Msx1_Lhx9_Barx1_Rax_Dlx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx1
|
ENSRNOG00000006876 | msh homeobox 1 |
|
Lhx9
|
ENSRNOG00000010357 | LIM homeobox 9 |
|
Barx1
|
ENSRNOG00000016915 | BARX homeobox 1 |
|
Rax
|
ENSRNOG00000016944 | retina and anterior neural fold homeobox |
|
Dlx6
|
ENSRNOG00000010822 | distal-less homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx6 | rn6_v1_chr4_+_32373641_32373641 | -0.54 | 3.9e-25 | Click! |
| Lhx9 | rn6_v1_chr13_-_55878094_55878094 | -0.47 | 9.1e-19 | Click! |
| Msx1 | rn6_v1_chr14_+_77712240_77712240 | -0.37 | 5.7e-12 | Click! |
| Rax | rn6_v1_chr18_-_61642056_61642056 | 0.17 | 2.0e-03 | Click! |
| Barx1 | rn6_v1_chr17_-_14828329_14828329 | 0.06 | 3.0e-01 | Click! |
Activity profile of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
Sorted Z-values of Msx1_Lhx9_Barx1_Rax_Dlx6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_5709236 | 16.84 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr7_-_143497108 | 10.33 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr10_-_87468711 | 9.97 |
ENSRNOT00000039983
|
Krtap3-3
|
keratin associated protein 3-3 |
| chr10_-_87232723 | 9.29 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr7_-_107391184 | 8.68 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr3_-_165537940 | 7.91 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr6_-_36876805 | 7.46 |
ENSRNOT00000006482
|
Msgn1
|
mesogenin 1 |
| chr6_-_128149220 | 7.23 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr10_-_87521514 | 6.67 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr7_-_143228060 | 6.07 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr16_+_22361998 | 6.05 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr8_+_47674321 | 5.78 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr10_-_87286387 | 5.51 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr16_+_2634603 | 5.31 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr7_-_73130740 | 4.57 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr12_-_45801842 | 4.02 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr14_+_59611434 | 3.76 |
ENSRNOT00000065366
|
Cckar
|
cholecystokinin A receptor |
| chr2_-_192027225 | 3.69 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr10_-_84920886 | 3.67 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chr1_-_127337882 | 3.64 |
ENSRNOT00000085158
|
Aldh1a3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr14_-_21252538 | 3.43 |
ENSRNOT00000005003
|
Ambn
|
ameloblastin |
| chr12_-_47142852 | 3.17 |
ENSRNOT00000001591
|
Pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr20_-_3728844 | 3.12 |
ENSRNOT00000074958
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 |
| chr1_+_105284753 | 3.06 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr6_+_8886591 | 2.97 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr7_+_143122269 | 2.81 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr14_+_23330933 | 2.51 |
ENSRNOT00000088552
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr4_+_147832136 | 2.51 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr1_+_279633671 | 2.42 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr10_+_53740841 | 2.32 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr4_+_61924013 | 2.32 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr7_+_41475163 | 2.31 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr7_+_133856101 | 2.29 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr20_+_44680449 | 2.24 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr7_-_69982592 | 2.23 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr17_+_25082056 | 2.22 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr3_+_9403840 | 2.21 |
ENSRNOT00000071380
|
NEWGENE_735020
|
pyroglutamylated RFamide peptide |
| chr2_-_33025271 | 2.20 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chrM_+_7006 | 2.15 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr3_-_90751055 | 2.10 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr7_-_49732764 | 2.04 |
ENSRNOT00000006453
|
Myf5
|
myogenic factor 5 |
| chr2_-_138833933 | 2.01 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr11_-_60679555 | 1.99 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr14_+_76732650 | 1.97 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr10_+_95770154 | 1.95 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr10_-_88172910 | 1.89 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr4_+_119225040 | 1.87 |
ENSRNOT00000012365
|
Bmp10
|
bone morphogenetic protein 10 |
| chr11_-_29710849 | 1.84 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr18_+_81694808 | 1.83 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr12_+_24978483 | 1.78 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr1_-_101095594 | 1.71 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr13_-_73056765 | 1.68 |
ENSRNOT00000000049
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr2_+_42829413 | 1.59 |
ENSRNOT00000065528
|
Actbl2
|
actin, beta-like 2 |
| chr15_+_44441856 | 1.56 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr4_-_172063391 | 1.56 |
ENSRNOT00000010158
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr2_+_210880777 | 1.51 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr6_-_125723732 | 1.49 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr20_-_30947484 | 1.44 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr6_-_21600451 | 1.42 |
ENSRNOT00000047674
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr2_-_61692487 | 1.37 |
ENSRNOT00000078544
|
LOC499541
|
LRRGT00045 |
| chr3_+_122544788 | 1.37 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr1_-_101819478 | 1.36 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr6_-_77508585 | 1.32 |
ENSRNOT00000011562
|
Nkx2-8
|
NK2 homeobox 8 |
| chr11_+_45751812 | 1.31 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr10_-_87503591 | 1.30 |
ENSRNOT00000037980
|
Krtap1-1
|
keratin associated protein 1-1 |
| chr4_-_168517177 | 1.27 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr14_+_23405717 | 1.22 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr15_-_95514259 | 1.20 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr11_+_47924970 | 1.20 |
ENSRNOT00000060577
|
Zpld1
|
zona pellucida-like domain containing 1 |
| chr7_-_126382449 | 1.18 |
ENSRNOT00000085540
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chr11_+_28780446 | 1.18 |
ENSRNOT00000072546
|
Krtap15-1
|
keratin associated protein 15-1 |
| chrM_+_8599 | 1.05 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr1_-_134871167 | 1.05 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_40137390 | 1.01 |
ENSRNOT00000042717
|
Panx3
|
pannexin 3 |
| chr2_+_240642847 | 0.99 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_-_122329443 | 0.91 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr16_+_10267482 | 0.90 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr4_+_1658278 | 0.90 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr5_+_50381244 | 0.88 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr8_+_71914867 | 0.83 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr4_+_158088505 | 0.75 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr8_-_63291966 | 0.74 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr3_+_77047466 | 0.72 |
ENSRNOT00000089424
|
Olr648
|
olfactory receptor 648 |
| chr3_+_56766475 | 0.69 |
ENSRNOT00000078819
|
Sp5
|
Sp5 transcription factor |
| chr6_-_77421286 | 0.68 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr2_+_266315036 | 0.67 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr1_+_101178104 | 0.63 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr4_-_67610710 | 0.59 |
ENSRNOT00000039067
|
Mrps33
|
mitochondrial ribosomal protein S33 |
| chr1_+_260093641 | 0.58 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr1_-_220938814 | 0.56 |
ENSRNOT00000028081
|
Ovol1
|
ovo like transcriptional repressor 1 |
| chr4_-_88684415 | 0.46 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr3_+_61620192 | 0.44 |
ENSRNOT00000065426
|
Hoxd9
|
homeo box D9 |
| chr13_+_49005405 | 0.44 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr4_+_1671269 | 0.42 |
ENSRNOT00000071452
|
Olr1251
|
olfactory receptor 1251 |
| chr2_-_32518643 | 0.40 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_+_96499520 | 0.39 |
ENSRNOT00000074692
|
AABR07016310.1
|
|
| chr17_-_53713408 | 0.38 |
ENSRNOT00000022445
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr5_+_25253010 | 0.36 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr4_-_80395502 | 0.33 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chrX_+_131381134 | 0.33 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr10_-_74679858 | 0.33 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr5_+_25725683 | 0.28 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr6_-_115616766 | 0.24 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr4_+_1591444 | 0.23 |
ENSRNOT00000078277
|
Olr1245
|
olfactory receptor 1245 |
| chr8_+_5790034 | 0.22 |
ENSRNOT00000061887
|
Mmp27
|
matrix metallopeptidase 27 |
| chr19_-_17399548 | 0.22 |
ENSRNOT00000075991
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr13_+_79378733 | 0.09 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr4_+_2055615 | 0.09 |
ENSRNOT00000007646
|
Rnf32
|
ring finger protein 32 |
| chr5_-_7941822 | 0.07 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr3_+_77431389 | 0.04 |
ENSRNOT00000042848
|
Olr660
|
olfactory receptor 660 |
| chr2_+_198755262 | 0.03 |
ENSRNOT00000028807
|
Rbm8a
|
RNA binding motif protein 8A |
| chr2_-_235249571 | 0.02 |
ENSRNOT00000093631
ENSRNOT00000085096 |
Rrh
|
retinal pigment epithelium derived rhodopsin homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx1_Lhx9_Barx1_Rax_Dlx6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 1.5 | 3.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.2 | 3.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.2 | 6.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.9 | 3.8 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.8 | 5.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.7 | 10.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.6 | 3.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.6 | 7.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.6 | 1.8 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.5 | 7.5 | GO:0007379 | segment specification(GO:0007379) |
| 0.4 | 7.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.4 | 2.3 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.4 | 1.9 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.3 | 9.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.3 | 2.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.3 | 0.8 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.3 | 1.6 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.3 | 2.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 2.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 1.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 0.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 2.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.7 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.1 | 1.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 5.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.7 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 2.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.7 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.6 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 2.2 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.1 | 0.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.9 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 | 5.8 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 1.4 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 3.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 6.1 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 3.7 | GO:0048144 | fibroblast proliferation(GO:0048144) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.6 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 2.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.0 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.0 | 2.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.3 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.8 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.7 | 3.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 3.2 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 39.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 1.6 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.3 | 2.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 14.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 3.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 2.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 7.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 6.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.8 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 2.0 | 6.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.9 | 3.4 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.7 | 2.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.6 | 1.8 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 2.3 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 1.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.4 | 3.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.3 | 1.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 3.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 2.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.3 | 5.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 1.9 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 1.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 3.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 7.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 3.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 10.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.7 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 7.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 36.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 1.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 9.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 3.8 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 8.2 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 2.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 10.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 7.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 4.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 10.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 9.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.3 | 2.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 1.6 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |