Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Mnx1_Lhx6_Lmx1a
Z-value: 0.52
Transcription factors associated with Mnx1_Lhx6_Lmx1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
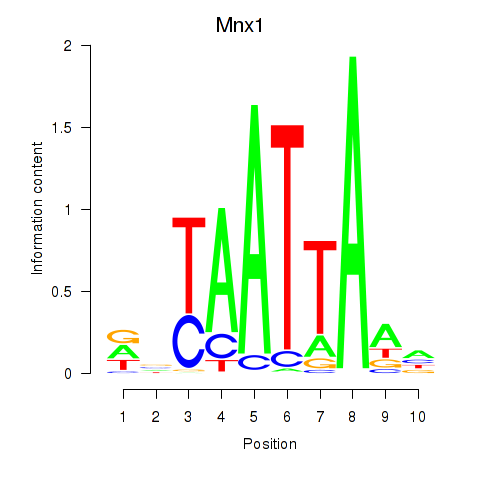
Mnx1
|
ENSRNOG00000046959 | motor neuron and pancreas homeobox 1 |
|
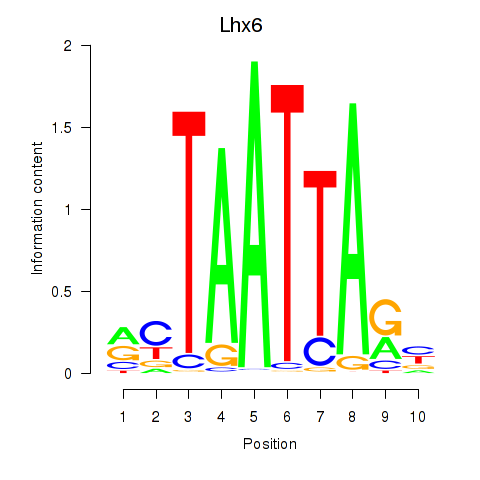
Lhx6
|
ENSRNOG00000005996 | LIM homeobox 6 |
|
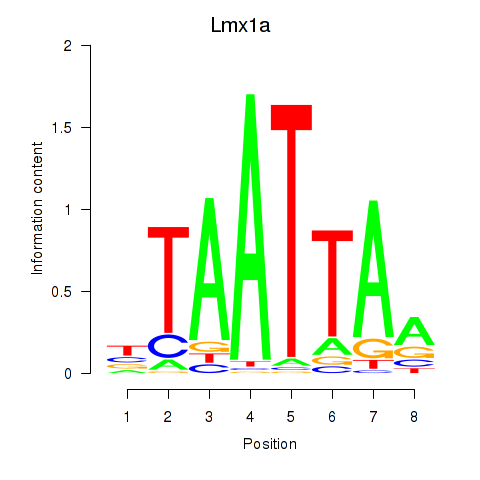
Lmx1a
|
ENSRNOG00000004642 | LIM homeobox transcription factor 1 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Lhx6 | rn6_v1_chr3_-_15433252_15433252 | -0.42 | 2.8e-15 | Click! |
| Lmx1a | rn6_v1_chr13_+_85918252_85918252 | -0.39 | 2.2e-13 | Click! |
| Mnx1 | rn6_v1_chr4_-_2381271_2381271 | -0.13 | 2.3e-02 | Click! |
Activity profile of Mnx1_Lhx6_Lmx1a motif
Sorted Z-values of Mnx1_Lhx6_Lmx1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_54466280 | 25.42 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr17_+_25082056 | 22.44 |
ENSRNOT00000037041
|
AABR07027339.1
|
|
| chr8_+_47674321 | 21.10 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr12_-_45801842 | 19.98 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr13_-_77821312 | 16.88 |
ENSRNOT00000082110
|
AABR07021544.1
|
|
| chr6_-_125723732 | 16.48 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr6_+_2216623 | 16.02 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr4_-_100252755 | 15.58 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr3_-_14229067 | 15.11 |
ENSRNOT00000025534
ENSRNOT00000092865 |
C5
|
complement C5 |
| chr8_+_106449321 | 14.08 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr4_+_22898527 | 13.64 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr8_+_71914867 | 13.57 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr1_+_279633671 | 13.00 |
ENSRNOT00000036012
ENSRNOT00000091669 |
Ccdc172
|
coiled-coil domain containing 172 |
| chr2_-_158156444 | 12.71 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr4_+_172119331 | 11.69 |
ENSRNOT00000010579
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr1_-_166912524 | 11.43 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr7_-_140291620 | 11.34 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr2_-_138833933 | 11.17 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr16_+_2634603 | 11.17 |
ENSRNOT00000019113
|
Hesx1
|
HESX homeobox 1 |
| chr7_-_73130740 | 10.61 |
ENSRNOT00000075584
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr2_-_158156150 | 10.19 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_-_71139267 | 9.73 |
ENSRNOT00000065232
|
RGD1561812
|
similar to Retinol dehydrogenase type II (RODH II) (29 k-protein) |
| chr7_-_69982592 | 9.70 |
ENSRNOT00000040010
|
RGD1564306
|
similar to developmental pluripotency associated 5 |
| chr3_-_57607683 | 9.54 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr3_-_90751055 | 9.53 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr5_+_50381244 | 9.44 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_-_101095594 | 9.43 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr14_+_76732650 | 9.23 |
ENSRNOT00000088197
|
Clnk
|
cytokine-dependent hematopoietic cell linker |
| chr14_+_91782354 | 9.09 |
ENSRNOT00000005902
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chrX_+_33443186 | 8.77 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr8_-_49109981 | 8.58 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr2_+_80269661 | 8.48 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr11_-_28478360 | 8.47 |
ENSRNOT00000032663
|
Cldn17
|
claudin 17 |
| chr16_+_10267482 | 8.28 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chr4_-_172063391 | 8.27 |
ENSRNOT00000010158
|
Slc15a5
|
solute carrier family 15, member 5 |
| chr1_+_88955440 | 8.02 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr10_-_87468711 | 7.81 |
ENSRNOT00000039983
|
Krtap3-3
|
keratin associated protein 3-3 |
| chr20_+_3176107 | 7.62 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr14_+_21177237 | 7.60 |
ENSRNOT00000004866
|
Jchain
|
immunoglobulin joining chain |
| chrX_+_156463953 | 7.37 |
ENSRNOT00000079889
|
Flna
|
filamin A |
| chr13_-_83457888 | 7.33 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr4_-_88684415 | 7.31 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr7_-_101138860 | 7.10 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr8_+_71514281 | 6.94 |
ENSRNOT00000022256
|
Ns5atp9
|
NS5A (hepatitis C virus) transactivated protein 9 |
| chr7_-_101138373 | 6.83 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr18_-_26656879 | 6.73 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr3_-_7498555 | 6.46 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr20_+_4357733 | 6.33 |
ENSRNOT00000000509
|
Pbx2
|
PBX homeobox 2 |
| chr18_+_81694808 | 6.28 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chrM_+_7006 | 6.08 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr11_-_79703736 | 6.05 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_43022565 | 6.01 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr7_-_143497108 | 6.01 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr8_+_99632803 | 5.93 |
ENSRNOT00000087190
|
Plscr1
|
phospholipid scramblase 1 |
| chr1_-_19376301 | 5.75 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr1_-_104973648 | 5.63 |
ENSRNOT00000019739
|
Dbx1
|
developing brain homeobox 1 |
| chr20_+_44680449 | 5.60 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr1_+_88955135 | 5.42 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr5_-_115387377 | 5.41 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr10_+_95770154 | 5.40 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr4_+_68849033 | 5.31 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr4_-_80395502 | 5.27 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr14_-_8600512 | 5.27 |
ENSRNOT00000092537
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr8_+_104106740 | 5.19 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr10_+_61685645 | 5.15 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr12_-_47142852 | 5.10 |
ENSRNOT00000001591
|
Pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr17_-_39824299 | 4.95 |
ENSRNOT00000023412
|
Prl
|
prolactin |
| chr6_-_125723944 | 4.92 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr1_-_6903486 | 4.90 |
ENSRNOT00000086574
|
Utrn
|
utrophin |
| chr2_+_266315036 | 4.89 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr11_-_62451149 | 4.85 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_165537940 | 4.77 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr13_-_83425641 | 4.63 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr14_+_23405717 | 4.55 |
ENSRNOT00000029805
|
Tmprss11c
|
transmembrane protease, serine 11C |
| chr10_-_88172910 | 4.53 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr8_+_117117430 | 4.52 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr13_-_90405591 | 4.52 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr4_+_6827429 | 4.43 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr7_+_121841855 | 4.37 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr3_+_110855000 | 4.31 |
ENSRNOT00000081613
|
Knl1
|
kinetochore scaffold 1 |
| chr6_-_95934296 | 4.24 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr16_+_50049828 | 4.21 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr4_-_170740274 | 4.18 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr20_+_3155652 | 4.14 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr1_-_263762785 | 4.05 |
ENSRNOT00000018221
|
Cpn1
|
carboxypeptidase N subunit 1 |
| chr1_+_253221812 | 4.04 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr9_-_79630452 | 4.00 |
ENSRNOT00000078125
ENSRNOT00000086044 ENSRNOT00000089283 |
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr9_+_88918433 | 3.97 |
ENSRNOT00000021730
|
Ccl20
|
C-C motif chemokine ligand 20 |
| chr8_+_22648323 | 3.95 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr6_-_128149220 | 3.93 |
ENSRNOT00000014204
|
Gsc
|
goosecoid homeobox |
| chr7_-_126382449 | 3.91 |
ENSRNOT00000085540
|
7530416G11Rik
|
RIKEN cDNA 7530416G11 gene |
| chrX_-_72034099 | 3.90 |
ENSRNOT00000004310
|
Ercc6l
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr14_-_44078897 | 3.89 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr7_-_2941122 | 3.88 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chr18_-_12640716 | 3.76 |
ENSRNOT00000020697
|
Klhl14
|
kelch-like family member 14 |
| chrX_-_32095867 | 3.72 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr2_-_61692487 | 3.71 |
ENSRNOT00000078544
|
LOC499541
|
LRRGT00045 |
| chr8_+_85259982 | 3.69 |
ENSRNOT00000010409
|
Elovl5
|
ELOVL fatty acid elongase 5 |
| chr16_-_85306366 | 3.66 |
ENSRNOT00000089650
|
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr2_-_31753528 | 3.63 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr17_-_80807181 | 3.60 |
ENSRNOT00000040052
ENSRNOT00000090064 |
Cubn
|
cubilin |
| chr4_+_158088505 | 3.60 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr1_+_22332090 | 3.58 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr5_+_165724027 | 3.50 |
ENSRNOT00000018000
|
Casz1
|
castor zinc finger 1 |
| chr1_+_107344904 | 3.46 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chrM_+_2740 | 3.45 |
ENSRNOT00000047550
|
Mt-nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr14_-_2032593 | 3.43 |
ENSRNOT00000000037
|
Fgfrl1
|
fibroblast growth factor receptor-like 1 |
| chr11_+_45751812 | 3.43 |
ENSRNOT00000079336
|
RGD1310935
|
similar to Dermal papilla derived protein 7 |
| chr1_-_276228574 | 3.38 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr10_-_87521514 | 3.37 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr5_-_147412705 | 3.31 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr2_+_188478082 | 3.30 |
ENSRNOT00000085817
|
Clk2
|
CDC-like kinase 2 |
| chr4_+_82637411 | 3.29 |
ENSRNOT00000048225
|
LOC685406
|
LRRGT00062 |
| chr2_-_60657712 | 3.21 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr9_-_4978892 | 3.18 |
ENSRNOT00000015189
|
Sult1c3
|
sulfotransferase family 1C member 3 |
| chr2_+_208749996 | 3.16 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr1_+_260093641 | 3.13 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr11_-_32550539 | 3.13 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr6_-_77421286 | 3.12 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chrX_+_43626480 | 3.09 |
ENSRNOT00000068078
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr10_-_88000423 | 3.07 |
ENSRNOT00000076787
ENSRNOT00000046751 ENSRNOT00000091394 |
Krt32
|
keratin 32 |
| chr17_-_57394985 | 3.05 |
ENSRNOT00000019968
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr9_-_14550625 | 3.04 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr11_-_70322690 | 3.03 |
ENSRNOT00000002443
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr5_-_102743417 | 3.02 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr4_-_103569159 | 2.93 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr13_+_92264231 | 2.91 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr4_-_21920651 | 2.91 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr1_+_13595295 | 2.88 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr4_+_31229913 | 2.87 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr8_+_48718329 | 2.86 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr2_-_88553086 | 2.83 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr10_-_65502936 | 2.83 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr2_+_243577082 | 2.79 |
ENSRNOT00000016556
|
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr5_+_25253010 | 2.76 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chrM_+_9451 | 2.75 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrM_+_8599 | 2.74 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr1_-_252461461 | 2.73 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr11_-_71284939 | 2.68 |
ENSRNOT00000002421
|
AABR07034428.1
|
|
| chr3_+_73302571 | 2.68 |
ENSRNOT00000085420
|
Olr468
|
olfactory receptor 468 |
| chr2_-_238369692 | 2.68 |
ENSRNOT00000041583
|
Arhgef38
|
Rho guanine nucleotide exchange factor 38 |
| chr4_+_147832136 | 2.60 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr17_+_9596957 | 2.58 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr14_+_23330933 | 2.54 |
ENSRNOT00000088552
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr5_+_25725683 | 2.54 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr8_+_57983556 | 2.45 |
ENSRNOT00000009562
|
RGD1311251
|
similar to RIKEN cDNA 4930550C14 |
| chr4_-_10995792 | 2.40 |
ENSRNOT00000078733
|
AABR07059243.1
|
|
| chrM_+_7919 | 2.39 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr19_+_53055745 | 2.36 |
ENSRNOT00000074430
|
Foxl1
|
forkhead box L1 |
| chr6_-_108660063 | 2.35 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr14_-_84189266 | 2.35 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr1_-_220938814 | 2.34 |
ENSRNOT00000028081
|
Ovol1
|
ovo like transcriptional repressor 1 |
| chr1_-_15374850 | 2.33 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr1_-_227932603 | 2.29 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr2_-_236431596 | 2.26 |
ENSRNOT00000055570
ENSRNOT00000089674 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr1_+_185210922 | 2.24 |
ENSRNOT00000055120
|
Pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chrM_+_11736 | 2.24 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_+_186776644 | 2.23 |
ENSRNOT00000046778
|
Fcrl3
|
Fc receptor-like 3 |
| chr1_-_101819478 | 2.23 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr16_+_4469468 | 2.19 |
ENSRNOT00000021164
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr8_-_53816447 | 2.19 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr16_+_29674793 | 2.19 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr2_+_208750356 | 2.18 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr10_+_105630810 | 2.17 |
ENSRNOT00000064904
|
Prcd
|
photoreceptor disc component |
| chr2_+_226563050 | 2.14 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr9_+_47536824 | 2.14 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr5_+_133819726 | 2.13 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chrM_+_10160 | 2.12 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr17_-_78812111 | 2.11 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr20_+_42966140 | 2.10 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr19_+_53044379 | 2.09 |
ENSRNOT00000072369
|
Foxc2
|
forkhead box C2 |
| chr2_+_252090669 | 2.09 |
ENSRNOT00000020656
|
Lpar3
|
lysophosphatidic acid receptor 3 |
| chr13_+_79378733 | 2.09 |
ENSRNOT00000039221
|
Tnfsf18
|
tumor necrosis factor superfamily member 18 |
| chr14_-_43694584 | 2.08 |
ENSRNOT00000041866
|
AABR07072400.1
|
|
| chr9_-_117484262 | 2.04 |
ENSRNOT00000091931
|
AABR07068750.1
|
|
| chr1_+_280633938 | 2.01 |
ENSRNOT00000012564
|
Emx2
|
empty spiracles homeobox 2 |
| chrM_+_9870 | 2.00 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr3_-_8659102 | 1.97 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr7_+_38900593 | 1.95 |
ENSRNOT00000072166
|
Epyc
|
epiphycan |
| chr5_+_6373583 | 1.95 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr1_+_101554642 | 1.94 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr3_-_75576520 | 1.94 |
ENSRNOT00000083330
|
Olr567
|
olfactory receptor 567 |
| chr1_-_134871167 | 1.94 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr3_+_73366155 | 1.93 |
ENSRNOT00000044979
|
Olr473
|
olfactory receptor 473 |
| chr1_-_94404211 | 1.91 |
ENSRNOT00000019463
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr16_-_85305782 | 1.86 |
ENSRNOT00000067511
ENSRNOT00000076737 |
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr3_+_2591331 | 1.86 |
ENSRNOT00000017466
|
Sapcd2
|
suppressor APC domain containing 2 |
| chr1_+_128637049 | 1.85 |
ENSRNOT00000018639
|
Ttc23
|
tetratricopeptide repeat domain 23 |
| chr6_+_8886591 | 1.79 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr14_-_45859908 | 1.77 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr2_-_33025271 | 1.73 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr11_+_47924970 | 1.69 |
ENSRNOT00000060577
|
Zpld1
|
zona pellucida-like domain containing 1 |
| chr1_+_101599018 | 1.66 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chrX_+_105147534 | 1.66 |
ENSRNOT00000046288
|
Cenpi
|
centromere protein I |
| chr10_-_87564327 | 1.64 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr3_-_76135475 | 1.60 |
ENSRNOT00000007712
|
Olr604
|
olfactory receptor 604 |
| chr5_+_33097654 | 1.57 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr3_+_5709236 | 1.55 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr12_+_47590154 | 1.55 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr7_+_133856101 | 1.55 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnx1_Lhx6_Lmx1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 25.4 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 5.2 | 15.6 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 4.5 | 13.6 | GO:2000424 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 4.5 | 13.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.5 | 7.6 | GO:0032679 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 2.5 | 15.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 2.3 | 9.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) |
| 2.1 | 21.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 2.0 | 5.9 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 1.9 | 11.3 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 1.8 | 14.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.6 | 4.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 1.6 | 11.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 1.5 | 4.5 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 1.5 | 4.5 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 1.5 | 6.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 1.5 | 7.4 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 1.4 | 4.2 | GO:1905242 | response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 1.4 | 5.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 1.3 | 4.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.3 | 4.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 1.3 | 4.0 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 1.3 | 7.6 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.2 | 3.7 | GO:0015827 | angiotensin-mediated drinking behavior(GO:0003051) tryptophan transport(GO:0015827) positive regulation of gap junction assembly(GO:1903598) |
| 1.2 | 3.5 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) retinal rod cell differentiation(GO:0060221) |
| 1.1 | 5.3 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.0 | 11.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.0 | 3.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.8 | 5.9 | GO:2000371 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.8 | 4.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.8 | 2.4 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.8 | 3.1 | GO:0032919 | spermine acetylation(GO:0032919) |
| 0.7 | 2.2 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.7 | 3.7 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 11.0 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.7 | 2.2 | GO:0061346 | serotonergic neuron axon guidance(GO:0036515) Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) |
| 0.7 | 14.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.7 | 2.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.6 | 1.9 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.6 | 14.7 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.6 | 5.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.6 | 6.0 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.6 | 1.8 | GO:0014016 | proximal/distal axis specification(GO:0009946) neuroblast differentiation(GO:0014016) |
| 0.6 | 4.4 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.5 | 5.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.5 | 2.1 | GO:0072011 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.5 | 1.5 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.5 | 1.5 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.5 | 2.9 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.5 | 2.8 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.5 | 6.0 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.5 | 1.4 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.4 | 2.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.4 | 8.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.4 | 3.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.4 | 3.9 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.4 | 2.3 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 2.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.3 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 4.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.3 | 5.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.3 | 0.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.3 | 3.4 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 2.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.3 | 2.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.3 | 2.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 3.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 | 2.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 3.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.3 | 4.9 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 4.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 | 2.8 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.2 | 1.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 21.7 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.2 | 4.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 0.9 | GO:0035582 | growth plate cartilage chondrocyte proliferation(GO:0003419) sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 6.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 12.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 1.1 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 2.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 1.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.2 | 6.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 3.0 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 3.4 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.2 | 1.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.2 | 0.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 2.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 0.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.2 | 1.7 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 1.0 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.4 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 0.1 | 0.9 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 1.7 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 2.9 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 2.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 2.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.5 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 6.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 9.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 2.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.3 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 0.5 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.5 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 5.1 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 9.6 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 1.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.5 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 0.9 | GO:1902358 | oxalate transport(GO:0019532) sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.7 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 4.1 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.0 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 2.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.5 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 4.7 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.2 | GO:0044206 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) |
| 0.0 | 1.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 1.0 | GO:0042990 | regulation of transcription factor import into nucleus(GO:0042990) |
| 0.0 | 2.1 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.9 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.5 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.4 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 3.1 | GO:0042278 | purine nucleoside metabolic process(GO:0042278) |
| 0.0 | 0.0 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 4.7 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 0.4 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 40.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 5.2 | 15.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 4.3 | 21.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 2.5 | 7.6 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 1.9 | 7.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.5 | 4.5 | GO:0060187 | cell pole(GO:0060187) |
| 1.2 | 3.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.9 | 4.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.8 | 5.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.8 | 3.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.7 | 18.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.7 | 2.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.7 | 4.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.6 | 9.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.6 | 3.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 8.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.4 | 1.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.4 | 1.5 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.4 | 6.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 1.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 3.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.3 | 2.9 | GO:0008091 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) cuticular plate(GO:0032437) |
| 0.3 | 3.9 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 12.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 2.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 16.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 1.4 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 21.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 2.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 7.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 12.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 4.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 2.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 9.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.2 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 3.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 17.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 7.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 4.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 7.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 10.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 10.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 3.5 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 4.5 | 13.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 2.4 | 17.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.9 | 11.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 1.8 | 12.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.5 | 7.6 | GO:0019862 | IgA binding(GO:0019862) |
| 1.5 | 8.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.3 | 5.3 | GO:0016160 | amylase activity(GO:0016160) |
| 1.3 | 15.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.3 | 3.9 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 1.3 | 11.4 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 1.2 | 6.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.1 | 14.1 | GO:0019841 | retinol binding(GO:0019841) |
| 1.0 | 3.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 1.0 | 21.1 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.0 | 4.0 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.0 | 4.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.9 | 2.8 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.9 | 3.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.9 | 5.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.8 | 5.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.8 | 3.2 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 0.8 | 3.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 6.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.7 | 2.2 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.7 | 16.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.6 | 1.9 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.6 | 2.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.6 | 3.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.6 | 1.7 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.5 | 1.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.5 | 5.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.5 | 4.0 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.5 | 12.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 3.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 5.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 2.9 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.4 | 1.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.4 | 7.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 2.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.4 | 3.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 1.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 2.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 1.3 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 2.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 0.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.3 | 7.5 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.3 | 2.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 1.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.3 | 3.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 4.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 30.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 13.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 13.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 1.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.2 | 2.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 4.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 8.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 4.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.6 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 7.6 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 12.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 11.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 4.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.6 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 9.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.8 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 9.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 9.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 5.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 15.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 4.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 14.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 3.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 4.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 5.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.3 | 8.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 9.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 11.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 9.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 3.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 24.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 5.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 3.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 40.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.0 | 13.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.9 | 9.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 15.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.5 | 11.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.5 | 4.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 17.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.5 | 4.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 14.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.4 | 11.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 3.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 2.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 8.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 3.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 2.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 3.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 4.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 3.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 6.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 5.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 9.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |