Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
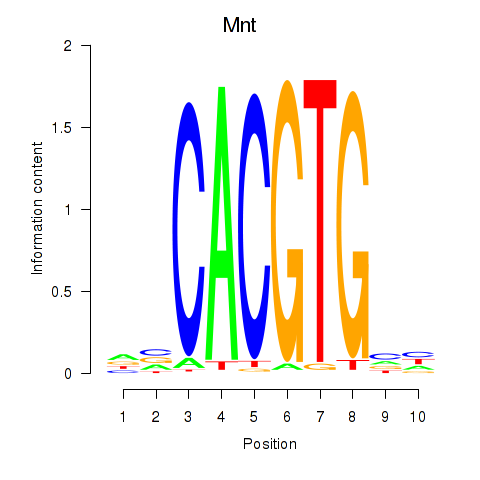
Results for Mnt
Z-value: 0.52
Transcription factors associated with Mnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mnt
|
ENSRNOG00000002894 | MAX network transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnt | rn6_v1_chr10_+_61685645_61685645 | -0.19 | 6.3e-04 | Click! |
Activity profile of Mnt motif
Sorted Z-values of Mnt motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_152681101 | 13.44 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr8_-_130127392 | 12.65 |
ENSRNOT00000026159
|
Cck
|
cholecystokinin |
| chr10_-_14937336 | 11.39 |
ENSRNOT00000025494
|
Sox8
|
SRY box 8 |
| chr9_-_82419288 | 10.62 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr4_+_33638709 | 9.93 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr5_+_152680407 | 9.14 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr19_+_55669626 | 7.60 |
ENSRNOT00000033352
|
Cdh15
|
cadherin 15 |
| chr8_+_22050222 | 7.48 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr1_+_203971152 | 7.34 |
ENSRNOT00000075540
|
Gpr26
|
G protein-coupled receptor 26 |
| chr1_+_102849889 | 7.32 |
ENSRNOT00000066791
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr7_-_13751271 | 7.23 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chr7_+_128500011 | 6.93 |
ENSRNOT00000074625
|
Fam19a5
|
family with sequence similarity 19 member A5, C-C motif chemokine like |
| chr2_+_243820661 | 6.69 |
ENSRNOT00000090526
|
Eif4e
|
eukaryotic translation initiation factor 4E |
| chr8_+_22402890 | 6.59 |
ENSRNOT00000079230
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr17_-_66397653 | 6.56 |
ENSRNOT00000024098
|
Actn2
|
actinin alpha 2 |
| chr13_-_111765944 | 6.55 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr7_-_140483693 | 6.48 |
ENSRNOT00000089060
|
Ddn
|
dendrin |
| chr3_-_66417741 | 6.39 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chrX_+_25016401 | 6.37 |
ENSRNOT00000059270
|
Clcn4
|
chloride voltage-gated channel 4 |
| chrX_-_138972684 | 6.15 |
ENSRNOT00000040165
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr18_+_16650806 | 5.98 |
ENSRNOT00000093679
ENSRNOT00000041961 ENSRNOT00000093140 |
Fhod3
|
formin homology 2 domain containing 3 |
| chr3_-_61494778 | 5.92 |
ENSRNOT00000068018
|
Lnpk
|
lunapark, ER junction formation factor |
| chrX_-_19134869 | 5.82 |
ENSRNOT00000004235
|
RragB
|
Ras-related GTP binding B |
| chrX_-_19369746 | 5.81 |
ENSRNOT00000075249
|
LOC108348096
|
ras-related GTP-binding protein B |
| chr20_-_5533600 | 5.80 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr4_+_175729726 | 5.67 |
ENSRNOT00000013230
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr10_+_55940533 | 5.61 |
ENSRNOT00000012061
|
RGD1563441
|
similar to RIKEN cDNA A030009H04 |
| chr1_-_100980340 | 5.60 |
ENSRNOT00000064272
|
Prmt1
|
protein arginine methyltransferase 1 |
| chr13_+_84474319 | 5.49 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr19_+_14653198 | 5.30 |
ENSRNOT00000020017
|
Rasd2
|
RASD family, member 2 |
| chr5_-_164844586 | 5.20 |
ENSRNOT00000011287
|
Clcn6
|
chloride voltage-gated channel 6 |
| chr3_-_161299024 | 5.11 |
ENSRNOT00000021216
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr10_-_4644570 | 4.81 |
ENSRNOT00000088279
|
Snn
|
stannin |
| chr7_+_143897014 | 4.81 |
ENSRNOT00000017314
|
Espl1
|
extra spindle pole bodies like 1, separase |
| chr2_+_115337439 | 4.77 |
ENSRNOT00000015779
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr7_+_77966722 | 4.69 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr6_+_41917132 | 4.64 |
ENSRNOT00000071387
|
Ntsr2
|
neurotensin receptor 2 |
| chr5_-_150520884 | 4.61 |
ENSRNOT00000085482
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr19_+_52086325 | 4.60 |
ENSRNOT00000020341
|
Necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr9_+_29984077 | 4.58 |
ENSRNOT00000075592
|
B3gat2
|
beta-1,3-glucuronyltransferase 2 |
| chr3_-_92290919 | 4.56 |
ENSRNOT00000007077
|
Fjx1
|
four jointed box 1 |
| chr10_-_37455022 | 4.49 |
ENSRNOT00000016742
|
LOC103694902
|
ubiquitin-conjugating enzyme E2 B |
| chr8_+_39305128 | 4.47 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr15_-_45376324 | 4.38 |
ENSRNOT00000076969
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr15_-_45376476 | 4.35 |
ENSRNOT00000065838
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr2_+_264704769 | 4.34 |
ENSRNOT00000012667
|
Depdc1
|
DEP domain containing 1 |
| chrX_-_29648359 | 4.32 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr8_-_36760742 | 4.31 |
ENSRNOT00000017307
|
Ddx25
|
DEAD-box helicase 25 |
| chr18_+_53915807 | 4.30 |
ENSRNOT00000026543
|
Adamts19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19 |
| chr14_+_4362717 | 4.21 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr16_+_49462889 | 4.20 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr2_+_34186091 | 4.14 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr3_+_122803772 | 4.13 |
ENSRNOT00000009564
|
Nop56
|
NOP56 ribonucleoprotein |
| chrX_+_62363757 | 4.09 |
ENSRNOT00000091240
|
Arx
|
aristaless related homeobox |
| chr1_-_198382614 | 4.09 |
ENSRNOT00000055016
|
Asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr6_+_26797126 | 4.06 |
ENSRNOT00000010586
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr14_+_38612408 | 3.90 |
ENSRNOT00000059516
|
Commd8
|
COMM domain containing 8 |
| chr7_-_70467453 | 3.90 |
ENSRNOT00000052288
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr3_+_103726238 | 3.86 |
ENSRNOT00000006776
|
Lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chrX_-_38196060 | 3.84 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr9_-_32019205 | 3.82 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr20_-_5533448 | 3.80 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr9_+_12633990 | 3.76 |
ENSRNOT00000066517
ENSRNOT00000077532 |
Dazl
|
deleted in azoospermia-like |
| chr8_-_91338843 | 3.73 |
ENSRNOT00000012999
|
Elovl4
|
ELOVL fatty acid elongase 4 |
| chr10_-_89958805 | 3.71 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr14_+_33580541 | 3.66 |
ENSRNOT00000002905
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr9_+_82345719 | 3.65 |
ENSRNOT00000057330
ENSRNOT00000078260 |
Fam134a
|
family with sequence similarity 134, member A |
| chr12_+_43940929 | 3.63 |
ENSRNOT00000001486
|
Rnft2
|
ring finger protein, transmembrane 2 |
| chr16_-_81706664 | 3.62 |
ENSRNOT00000026580
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr4_-_157679962 | 3.61 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_198706852 | 3.61 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr5_+_165405168 | 3.60 |
ENSRNOT00000014934
|
Srm
|
spermidine synthase |
| chr1_+_40982862 | 3.60 |
ENSRNOT00000026367
ENSRNOT00000092953 |
Armt1
|
acidic residue methyltransferase 1 |
| chr7_-_82059247 | 3.52 |
ENSRNOT00000087177
|
Rspo2
|
R-spondin 2 |
| chr1_-_198232344 | 3.49 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chrX_-_64702441 | 3.48 |
ENSRNOT00000051132
|
Amer1
|
APC membrane recruitment protein 1 |
| chr19_+_26151900 | 3.48 |
ENSRNOT00000005571
|
Tnpo2
|
transportin 2 |
| chr10_-_81587858 | 3.45 |
ENSRNOT00000003582
|
Utp18
|
UTP18 small subunit processome component |
| chr1_-_104202591 | 3.45 |
ENSRNOT00000035512
|
E2f8
|
E2F transcription factor 8 |
| chr2_-_211017778 | 3.45 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr11_+_80255790 | 3.39 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr3_+_72226613 | 3.38 |
ENSRNOT00000010619
|
Timm10
|
translocase of inner mitochondrial membrane 10 |
| chr12_+_23544287 | 3.33 |
ENSRNOT00000001938
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr6_-_127248372 | 3.31 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr6_-_46631983 | 3.31 |
ENSRNOT00000045963
|
Sox11
|
SRY box 11 |
| chr18_-_4294136 | 3.27 |
ENSRNOT00000091909
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr9_-_11027506 | 3.21 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr1_+_100393303 | 3.20 |
ENSRNOT00000026251
|
Syt3
|
synaptotagmin 3 |
| chr2_-_188561267 | 3.20 |
ENSRNOT00000089781
ENSRNOT00000092093 |
Trim46
|
tripartite motif-containing 46 |
| chr1_-_265899958 | 3.18 |
ENSRNOT00000026013
|
Pitx3
|
paired-like homeodomain 3 |
| chr2_+_187893875 | 3.17 |
ENSRNOT00000093014
|
Mex3a
|
mex-3 RNA binding family member A |
| chr1_-_259089632 | 3.17 |
ENSRNOT00000020940
ENSRNOT00000091297 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_+_157165888 | 3.16 |
ENSRNOT00000043029
|
Pla2g2c
|
phospholipase A2, group IIC |
| chr1_-_259674425 | 3.16 |
ENSRNOT00000091116
|
LOC108348083
|
delta-1-pyrroline-5-carboxylate synthase |
| chr1_-_261140389 | 3.15 |
ENSRNOT00000073193
|
Rrp12
|
ribosomal RNA processing 12 homolog |
| chr3_-_123119460 | 3.13 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chrX_+_62363953 | 3.11 |
ENSRNOT00000083362
|
Arx
|
aristaless related homeobox |
| chr11_+_81972219 | 3.10 |
ENSRNOT00000002452
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr1_+_142679345 | 3.10 |
ENSRNOT00000034267
|
Zscan2
|
zinc finger and SCAN domain containing 2 |
| chr2_+_189400696 | 3.07 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr20_-_5723902 | 3.06 |
ENSRNOT00000036871
|
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr1_+_198383201 | 3.05 |
ENSRNOT00000037405
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr13_-_42263024 | 3.03 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr10_+_74436208 | 3.02 |
ENSRNOT00000090921
ENSRNOT00000091163 |
Trim37
|
tripartite motif-containing 37 |
| chr11_+_34598492 | 3.01 |
ENSRNOT00000065600
|
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr5_-_136748980 | 3.01 |
ENSRNOT00000026844
|
Ipo13
|
importin 13 |
| chr17_+_54280851 | 3.01 |
ENSRNOT00000024022
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr5_-_135677432 | 2.98 |
ENSRNOT00000024393
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr3_+_155297566 | 2.98 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr10_-_89959176 | 2.97 |
ENSRNOT00000028264
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr7_+_11521998 | 2.96 |
ENSRNOT00000026960
|
Sgta
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr18_+_51785111 | 2.95 |
ENSRNOT00000019351
|
Lmnb1
|
lamin B1 |
| chr1_-_101457126 | 2.94 |
ENSRNOT00000087061
ENSRNOT00000028328 |
Bax
|
BCL2 associated X, apoptosis regulator |
| chr1_-_255376833 | 2.91 |
ENSRNOT00000024941
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr11_+_30550141 | 2.89 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr1_-_226255886 | 2.89 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr7_+_108613739 | 2.88 |
ENSRNOT00000007564
|
Phf20l1
|
PHD finger protein 20-like 1 |
| chr10_+_89236256 | 2.83 |
ENSRNOT00000027957
|
Psme3
|
proteasome activator subunit 3 |
| chr10_+_74436534 | 2.80 |
ENSRNOT00000008298
|
Trim37
|
tripartite motif-containing 37 |
| chr8_+_63379087 | 2.78 |
ENSRNOT00000012091
ENSRNOT00000012295 |
Nptn
|
neuroplastin |
| chr14_-_42221225 | 2.76 |
ENSRNOT00000036103
|
Shisa3
|
shisa family member 3 |
| chr7_-_83701447 | 2.70 |
ENSRNOT00000088893
|
AABR07057675.1
|
|
| chr4_+_120411991 | 2.67 |
ENSRNOT00000090875
|
Ruvbl1
|
RuvB-like AAA ATPase 1 |
| chr5_-_152458023 | 2.67 |
ENSRNOT00000031225
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr3_-_12944494 | 2.66 |
ENSRNOT00000023172
|
Mvb12b
|
multivesicular body subunit 12B |
| chr17_-_46794845 | 2.63 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr20_+_31265483 | 2.63 |
ENSRNOT00000079584
ENSRNOT00000000674 |
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr1_+_78800754 | 2.62 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr6_-_75763185 | 2.62 |
ENSRNOT00000072589
|
Cfl2
|
cofilin 2 |
| chr8_+_64489942 | 2.60 |
ENSRNOT00000083666
|
Pkm
|
pyruvate kinase, muscle |
| chr1_+_85112834 | 2.60 |
ENSRNOT00000026107
|
Dyrk1b
|
dual specificity tyrosine phosphorylation regulated kinase 1B |
| chr20_-_10386663 | 2.58 |
ENSRNOT00000045275
ENSRNOT00000042432 |
Cbs
|
cystathionine beta synthase |
| chr7_+_53878610 | 2.57 |
ENSRNOT00000091910
|
Osbpl8
|
oxysterol binding protein-like 8 |
| chr1_+_222229835 | 2.57 |
ENSRNOT00000051749
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr7_+_144865608 | 2.54 |
ENSRNOT00000091596
ENSRNOT00000055285 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr3_+_138504214 | 2.54 |
ENSRNOT00000091529
|
Kat14
|
lysine acetyltransferase 14 |
| chr7_-_27214236 | 2.54 |
ENSRNOT00000034213
|
Tdg
|
thymine-DNA glycosylase |
| chr5_+_150725654 | 2.54 |
ENSRNOT00000089852
ENSRNOT00000017740 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr3_+_173953869 | 2.50 |
ENSRNOT00000091212
|
Fam217b
|
family with sequence similarity 217, member B |
| chr18_+_14756684 | 2.47 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_-_78333321 | 2.42 |
ENSRNOT00000020402
|
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr10_+_70262361 | 2.42 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_+_207108552 | 2.36 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr5_+_39251337 | 2.34 |
ENSRNOT00000010073
|
Ndufaf4
|
NADH:ubiquinone oxidoreductase complex assembly factor 4 |
| chr18_+_3597240 | 2.30 |
ENSRNOT00000017045
|
RGD1311805
|
similar to RIKEN cDNA 2400010D15 |
| chr7_-_121058029 | 2.30 |
ENSRNOT00000068033
|
Cbx6
|
chromobox 6 |
| chr1_+_77994203 | 2.29 |
ENSRNOT00000002044
|
Napa
|
NSF attachment protein alpha |
| chr5_-_136721379 | 2.28 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr6_-_26314844 | 2.26 |
ENSRNOT00000006658
|
Zfp512
|
zinc finger protein 512 |
| chr5_-_150323063 | 2.25 |
ENSRNOT00000014084
|
Oprd1
|
opioid receptor, delta 1 |
| chr7_-_27213651 | 2.24 |
ENSRNOT00000077041
ENSRNOT00000076468 |
Tdg
|
thymine-DNA glycosylase |
| chr5_+_140979435 | 2.23 |
ENSRNOT00000056592
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr5_-_165259224 | 2.23 |
ENSRNOT00000012692
|
Ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr3_-_3295101 | 2.21 |
ENSRNOT00000051729
|
Sohlh1
|
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr6_-_137733026 | 2.21 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr6_+_110968061 | 2.20 |
ENSRNOT00000014980
|
Cipc
|
CLOCK-interacting pacemaker |
| chr3_+_113415774 | 2.18 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr10_-_56850085 | 2.17 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr9_-_20621051 | 2.17 |
ENSRNOT00000015918
|
Tnfrsf21
|
TNF receptor superfamily member 21 |
| chr17_+_16333415 | 2.14 |
ENSRNOT00000060550
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr16_-_75855745 | 2.14 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr17_-_9695292 | 2.13 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr3_+_138770574 | 2.11 |
ENSRNOT00000012510
ENSRNOT00000066986 |
Dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chrX_-_32355296 | 2.10 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_+_56546710 | 2.10 |
ENSRNOT00000023003
|
Ybx2
|
Y box binding protein 2 |
| chr15_-_88670349 | 2.08 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr3_-_163935617 | 2.08 |
ENSRNOT00000074023
|
Kcnb1
|
potassium voltage-gated channel subfamily B member 1 |
| chr9_+_15393330 | 2.06 |
ENSRNOT00000075630
|
Bysl
|
bystin-like |
| chr9_+_16612433 | 2.04 |
ENSRNOT00000023979
|
Klhdc3
|
kelch domain containing 3 |
| chr2_+_115678344 | 2.02 |
ENSRNOT00000036717
|
Slc7a14
|
solute carrier family 7, member 14 |
| chr1_-_170578693 | 2.02 |
ENSRNOT00000025651
|
Rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr7_+_144565392 | 2.01 |
ENSRNOT00000021589
|
Hoxc11
|
homeobox C11 |
| chr3_-_92242318 | 2.00 |
ENSRNOT00000007018
|
Trim44
|
tripartite motif-containing 44 |
| chr5_-_711033 | 1.99 |
ENSRNOT00000024199
|
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr16_-_20939545 | 1.99 |
ENSRNOT00000027457
|
Sugp2
|
SURP and G patch domain containing 2 |
| chr13_+_72804218 | 1.97 |
ENSRNOT00000035145
|
Stx6
|
syntaxin 6 |
| chr5_+_135574172 | 1.96 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr3_-_94686989 | 1.96 |
ENSRNOT00000016677
|
Depdc7
|
DEP domain containing 7 |
| chr20_-_43108198 | 1.95 |
ENSRNOT00000000742
|
Hdac2
|
histone deacetylase 2 |
| chr8_-_132790778 | 1.93 |
ENSRNOT00000008255
ENSRNOT00000091431 |
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr1_-_102849430 | 1.92 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chr1_-_24191908 | 1.91 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr1_-_84008293 | 1.91 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr10_-_83898527 | 1.90 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr10_-_90983928 | 1.89 |
ENSRNOT00000055176
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr10_-_13580821 | 1.86 |
ENSRNOT00000009735
|
Ntn3
|
netrin 3 |
| chr12_+_24367199 | 1.86 |
ENSRNOT00000001971
|
Fkbp6
|
FK506 binding protein 6 |
| chr10_-_88667345 | 1.85 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr5_+_120250616 | 1.84 |
ENSRNOT00000070967
|
Ak4
|
adenylate kinase 4 |
| chr16_+_20939655 | 1.83 |
ENSRNOT00000027550
|
Armc6
|
armadillo repeat containing 6 |
| chr3_-_10161989 | 1.83 |
ENSRNOT00000012312
|
Exosc2
|
exosome component 2 |
| chr7_+_72772440 | 1.83 |
ENSRNOT00000009989
|
Mtdh
|
metadherin |
| chr5_+_150459713 | 1.81 |
ENSRNOT00000081681
ENSRNOT00000074251 |
Taf12
|
TATA-box binding protein associated factor 12 |
| chr10_+_70134729 | 1.80 |
ENSRNOT00000076739
|
Lig3
|
DNA ligase 3 |
| chr13_-_101697684 | 1.80 |
ENSRNOT00000078834
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr10_-_71837851 | 1.79 |
ENSRNOT00000000258
|
Aatf
|
apoptosis antagonizing transcription factor |
| chr10_+_56610051 | 1.78 |
ENSRNOT00000024348
|
Dvl2
|
dishevelled segment polarity protein 2 |
| chr10_+_82375572 | 1.76 |
ENSRNOT00000004947
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr9_+_94425252 | 1.76 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr8_+_44990014 | 1.76 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 22.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 2.8 | 11.4 | GO:0072034 | retinal rod cell differentiation(GO:0060221) renal vesicle induction(GO:0072034) |
| 2.2 | 6.6 | GO:0086097 | actin filament uncapping(GO:0051695) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) positive regulation of endocytic recycling(GO:2001137) |
| 2.0 | 6.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 2.0 | 9.9 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 1.6 | 6.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.6 | 4.8 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 1.5 | 5.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 1.4 | 5.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 1.4 | 4.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 1.2 | 6.2 | GO:0042262 | DNA protection(GO:0042262) |
| 1.2 | 3.7 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 1.2 | 3.6 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.2 | 3.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.2 | 7.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.2 | 4.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 1.1 | 4.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.1 | 4.3 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 1.1 | 3.2 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 1.0 | 3.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 1.0 | 2.9 | GO:1902445 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 1.0 | 12.6 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 1.0 | 5.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.9 | 4.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.8 | 3.4 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.8 | 5.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.8 | 4.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.8 | 3.3 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.8 | 3.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.8 | 3.8 | GO:0061743 | motor learning(GO:0061743) |
| 0.7 | 3.7 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 2.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.7 | 2.8 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.7 | 2.7 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.7 | 2.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.7 | 3.9 | GO:0006449 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 0.7 | 2.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.6 | 2.6 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.6 | 3.2 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.6 | 3.2 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.6 | 2.5 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.6 | 6.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.6 | 0.6 | GO:1904585 | response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 0.6 | 1.8 | GO:0071038 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.6 | 0.6 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.6 | 1.8 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.6 | 3.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.6 | 2.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.5 | 2.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.5 | 2.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.5 | 4.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.5 | 3.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 5.7 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 1.5 | GO:1905204 | septum secundum development(GO:0003285) negative regulation of connective tissue replacement(GO:1905204) |
| 0.5 | 3.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 1.5 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.5 | 1.0 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.5 | 1.4 | GO:0015820 | leucine transport(GO:0015820) |
| 0.4 | 1.8 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.4 | 1.7 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.4 | 1.3 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.4 | 1.3 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) regulation of amacrine cell differentiation(GO:1902869) |
| 0.4 | 2.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.4 | 1.2 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.4 | 1.6 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.4 | 2.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.4 | 1.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 6.6 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.4 | 1.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.4 | 1.2 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.4 | 1.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 | 1.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.4 | 2.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.4 | 1.8 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.4 | 4.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 7.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.3 | 2.8 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.3 | 3.8 | GO:0007135 | meiosis II(GO:0007135) |
| 0.3 | 1.0 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 0.3 | 4.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.9 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.3 | 2.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.3 | 2.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 0.9 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.3 | 0.9 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.3 | 1.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.3 | 0.6 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.3 | 0.8 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.3 | 3.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 1.9 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.3 | 4.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.3 | 1.3 | GO:1904528 | positive regulation of microtubule binding(GO:1904528) |
| 0.3 | 1.8 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.3 | 13.2 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.3 | 2.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.3 | 2.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.3 | 4.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.3 | 2.0 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.3 | 4.8 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 3.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.7 | GO:0046295 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.2 | 2.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 4.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 1.3 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 | 1.1 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.2 | 1.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 1.5 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 1.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 2.0 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 | 1.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 0.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.2 | 1.5 | GO:0003360 | brainstem development(GO:0003360) |
| 0.2 | 1.5 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.2 | 2.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 10.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 2.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 3.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.2 | 4.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 0.7 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.2 | 0.5 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.2 | 1.0 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.2 | 1.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 9.7 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.2 | 1.3 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.2 | 1.9 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.2 | 1.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 | 7.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 | 0.8 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 3.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 1.7 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 1.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 2.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 5.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.7 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 4.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 6.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.7 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.1 | 1.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.3 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 2.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 1.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 4.6 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 4.0 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 2.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.1 | 2.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.7 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.1 | 2.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 3.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 3.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.4 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 1.7 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 0.4 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 3.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 6.0 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 1.7 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 1.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 1.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 1.6 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 1.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.8 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 3.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 4.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 2.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 4.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 2.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.0 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 3.4 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.6 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.1 | 4.3 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 0.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 0.5 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.4 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 1.0 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 3.0 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 4.6 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.3 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.0 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 3.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 2.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 1.2 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.4 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 3.0 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 2.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.1 | GO:1904031 | positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.8 | GO:1903205 | regulation of hydrogen peroxide-induced cell death(GO:1903205) |
| 0.0 | 1.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.2 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.9 | GO:0050482 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 1.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 4.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 4.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 1.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 3.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.4 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 1.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 2.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.3 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 2.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.0 | GO:0003264 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) regulation of secondary heart field cardioblast proliferation(GO:0003266) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 12.6 | GO:0043203 | axon hillock(GO:0043203) |
| 1.3 | 3.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.0 | 5.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 1.0 | 6.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.9 | 2.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.9 | 4.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.9 | 8.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.9 | 3.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.9 | 3.5 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.9 | 6.0 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.8 | 2.5 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.8 | 2.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.6 | 3.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.6 | 3.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.6 | 1.8 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 0.6 | 1.8 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.6 | 4.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.6 | 3.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.5 | 3.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.5 | 1.6 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.5 | 2.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.5 | 2.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 1.8 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.4 | 1.3 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.4 | 6.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 3.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 7.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 7.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 2.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 3.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.4 | 5.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 3.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 1.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 1.0 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 0.3 | 2.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 1.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 1.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.3 | 2.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 3.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 1.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 6.0 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 4.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 5.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 5.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 1.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 3.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 1.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.2 | 0.8 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 1.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 2.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 2.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 0.5 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 2.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 1.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 2.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 7.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 40.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.8 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 11.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 10.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 0.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.3 | GO:0097361 | MMXD complex(GO:0071817) CIA complex(GO:0097361) |
| 0.1 | 4.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 4.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 9.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 4.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 2.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 3.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 3.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 2.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 8.8 | GO:0036477 | somatodendritic compartment(GO:0036477) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 1.5 | 4.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.3 | 6.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 1.2 | 4.6 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 1.2 | 4.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.1 | 5.6 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 1.1 | 6.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 1.0 | 3.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.9 | 3.6 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.9 | 6.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.8 | 2.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 3.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.7 | 8.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.7 | 5.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.7 | 2.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.6 | 2.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 12.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.6 | 2.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.6 | 2.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.5 | 2.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 7.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.5 | 3.6 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.5 | 3.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 2.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.5 | 1.5 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.5 | 1.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.5 | 3.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.5 | 3.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.4 | 2.7 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.4 | 1.8 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.4 | 12.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 3.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 1.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.4 | 6.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.4 | 5.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 2.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 3.6 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.4 | 9.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.4 | 1.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.4 | 7.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.4 | 1.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.4 | 3.0 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.4 | 2.6 | GO:1904047 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.4 | 3.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 6.0 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.3 | 1.0 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 1.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.3 | 2.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 3.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.3 | 1.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 4.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.3 | 1.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.3 | 1.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 2.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.5 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 5.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 3.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 6.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 3.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.6 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 1.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 2.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.8 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 15.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 3.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.2 | 4.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 1.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 12.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 1.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 2.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 1.6 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.9 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 1.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 2.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.5 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 1.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 1.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 4.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.4 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 8.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 3.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 1.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 4.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 3.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 7.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 0.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 4.0 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.1 | 0.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 2.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.1 | 4.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 0.6 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 3.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 4.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 7.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 7.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 19.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 2.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 7.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.9 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 4.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.9 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 10.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 3.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.8 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 3.5 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 0.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.2 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 5.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 3.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.3 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 7.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 4.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 22.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 3.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 1.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 4.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 4.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 6.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 5.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 4.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 3.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 5.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.6 | 11.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 6.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 4.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 7.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 3.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 7.8 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.3 | 7.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 0.8 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.3 | 7.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 2.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 5.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.2 | 2.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 4.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 29.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 4.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 2.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 4.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 5.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 5.4 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 2.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.7 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 6.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.3 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.1 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.7 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 4.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 13.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 1.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.2 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |