Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
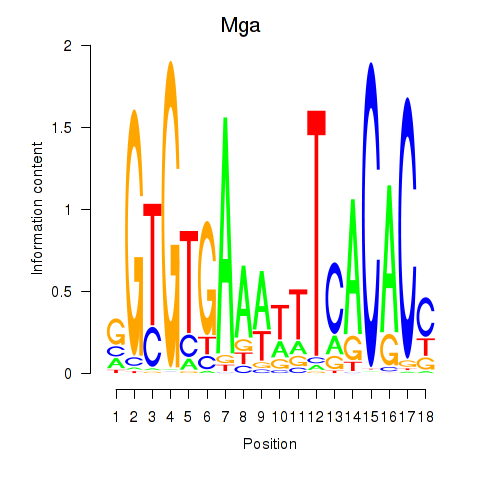
Results for Mga
Z-value: 0.77
Transcription factors associated with Mga
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mga
|
ENSRNOG00000006378 | MGA, MAX dimerization protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mga | rn6_v1_chr3_+_111699021_111699087 | 0.21 | 1.2e-04 | Click! |
Activity profile of Mga motif
Sorted Z-values of Mga motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_110232843 | 32.11 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr16_-_31301880 | 29.12 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr3_+_19772056 | 29.04 |
ENSRNOT00000044455
|
AABR07051708.1
|
|
| chr4_+_163162211 | 25.23 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr5_-_138239306 | 22.60 |
ENSRNOT00000039305
|
Ermap
|
erythroblast membrane-associated protein |
| chr7_-_12635943 | 20.18 |
ENSRNOT00000015029
|
Cfd
|
complement factor D |
| chr14_-_5859581 | 18.38 |
ENSRNOT00000052308
|
AABR07014241.1
|
|
| chr3_-_20695952 | 17.87 |
ENSRNOT00000072306
|
AABR07051746.1
|
|
| chrX_+_122507374 | 16.57 |
ENSRNOT00000032275
ENSRNOT00000080517 |
Dock11
|
dedicator of cytokinesis 11 |
| chr2_+_87418517 | 15.66 |
ENSRNOT00000048046
|
LOC100909879
|
tyrosine-protein phosphatase non-receptor type substrate 1-like |
| chr19_-_37952501 | 15.44 |
ENSRNOT00000026809
|
Dpep3
|
dipeptidase 3 |
| chr7_+_119820537 | 15.26 |
ENSRNOT00000077256
ENSRNOT00000056221 |
Cyth4
|
cytohesin 4 |
| chr15_-_28081465 | 15.02 |
ENSRNOT00000033739
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr8_-_50526843 | 14.96 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chr7_-_117680004 | 14.49 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr3_+_20007192 | 13.90 |
ENSRNOT00000075229
|
AABR07051716.1
|
|
| chr7_+_140758615 | 13.41 |
ENSRNOT00000089448
|
Troap
|
trophinin associated protein |
| chr8_+_5768811 | 13.32 |
ENSRNOT00000013936
|
Mmp8
|
matrix metallopeptidase 8 |
| chr3_-_46601409 | 13.04 |
ENSRNOT00000079261
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr10_-_57837602 | 12.40 |
ENSRNOT00000075185
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr8_-_114449956 | 12.36 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr20_-_3401273 | 11.31 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr14_-_84662143 | 10.84 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr3_+_17180411 | 10.75 |
ENSRNOT00000058260
|
AABR07051565.1
|
|
| chr10_+_105498728 | 10.53 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr20_-_7219548 | 10.46 |
ENSRNOT00000033907
|
Rps10
|
ribosomal protein S10 |
| chr1_+_252589785 | 10.34 |
ENSRNOT00000025928
|
Fas
|
Fas cell surface death receptor |
| chr4_-_78342863 | 10.25 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr1_-_101095594 | 10.25 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chrX_-_118513061 | 9.97 |
ENSRNOT00000043338
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr1_+_79754587 | 9.90 |
ENSRNOT00000083211
|
Micb
|
MHC class I polypeptide-related sequence B |
| chr9_+_82632230 | 9.84 |
ENSRNOT00000085165
|
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr2_+_186703541 | 9.83 |
ENSRNOT00000093342
|
Fcrl1
|
Fc receptor-like 1 |
| chr1_+_213040024 | 9.66 |
ENSRNOT00000050237
|
Olr300
|
olfactory receptor 300 |
| chr20_-_5618254 | 9.56 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr6_-_79306443 | 9.10 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr17_+_43632397 | 8.86 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr4_+_154391647 | 8.84 |
ENSRNOT00000081488
ENSRNOT00000079192 |
Mug1
|
murinoglobulin 1 |
| chr4_-_154855098 | 8.57 |
ENSRNOT00000041957
|
LOC297568
|
alpha-1-inhibitor III |
| chr6_-_127816055 | 8.54 |
ENSRNOT00000013175
|
Serpina3m
|
serine (or cysteine) proteinase inhibitor, clade A, member 3M |
| chr4_-_164306500 | 8.39 |
ENSRNOT00000080133
|
Ly49i5
|
Ly49 inhibitory receptor 5 |
| chr7_-_65275408 | 8.08 |
ENSRNOT00000065137
|
Hmga2
|
high mobility group AT-hook 2 |
| chr18_+_16590408 | 7.74 |
ENSRNOT00000093715
|
Mocos
|
molybdenum cofactor sulfurase |
| chr4_+_181481147 | 7.63 |
ENSRNOT00000002523
|
Klhl42
|
kelch-like family, member 42 |
| chr10_+_105500290 | 7.41 |
ENSRNOT00000079080
ENSRNOT00000083593 |
Sphk1
|
sphingosine kinase 1 |
| chr18_-_70924708 | 7.31 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr16_-_81434363 | 6.68 |
ENSRNOT00000080963
|
Rasa3
|
RAS p21 protein activator 3 |
| chr20_+_20519031 | 6.65 |
ENSRNOT00000000780
|
AABR07044783.1
|
|
| chr11_+_67555658 | 6.58 |
ENSRNOT00000039075
|
Csta
|
cystatin A (stefin A) |
| chr7_+_126096793 | 6.51 |
ENSRNOT00000047234
ENSRNOT00000088042 |
Fbln1
|
fibulin 1 |
| chr9_+_69497121 | 6.37 |
ENSRNOT00000042562
|
Nrp2
|
neuropilin 2 |
| chr7_+_120176530 | 6.29 |
ENSRNOT00000087548
|
Triobp
|
TRIO and F-actin binding protein |
| chr2_-_200762206 | 6.13 |
ENSRNOT00000068511
ENSRNOT00000086835 |
Hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr12_-_6879154 | 5.89 |
ENSRNOT00000001207
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr16_-_83132785 | 5.74 |
ENSRNOT00000043449
|
Arhgef7
|
Rho guanine nucleotide exchange factor 7 |
| chr3_+_72540538 | 5.56 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr8_-_119523716 | 5.45 |
ENSRNOT00000064581
|
Mlh1
|
mutL homolog 1 |
| chr8_-_119523964 | 5.18 |
ENSRNOT00000081718
|
Mlh1
|
mutL homolog 1 |
| chr8_-_132027006 | 5.17 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr5_+_69833225 | 5.09 |
ENSRNOT00000013759
|
Nipsnap3b
|
nipsnap homolog 3B |
| chrX_-_1786978 | 5.03 |
ENSRNOT00000011317
|
Rbm10
|
RNA binding motif protein 10 |
| chr1_+_222250995 | 4.92 |
ENSRNOT00000031458
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr9_+_82632384 | 4.56 |
ENSRNOT00000027064
|
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr18_+_24540659 | 4.54 |
ENSRNOT00000061074
|
Ammecr1l
|
AMMECR1 like |
| chr6_+_106496992 | 4.49 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr8_-_22929294 | 4.30 |
ENSRNOT00000015609
|
Rab3d
|
RAB3D, member RAS oncogene family |
| chr13_-_61306939 | 4.30 |
ENSRNOT00000086610
|
Rgs21
|
regulator of G-protein signaling 21 |
| chr20_-_14393793 | 4.29 |
ENSRNOT00000001764
|
Specc1l
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr9_+_15309127 | 4.24 |
ENSRNOT00000019177
|
Prickle4
|
prickle planar cell polarity protein 4 |
| chr6_+_12253788 | 3.96 |
ENSRNOT00000061675
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chrX_-_159206949 | 3.93 |
ENSRNOT00000071516
|
Map7d3
|
MAP7 domain containing 3 |
| chr1_+_221409271 | 3.88 |
ENSRNOT00000028462
|
Syvn1
|
synoviolin 1 |
| chr10_+_71008709 | 3.84 |
ENSRNOT00000055923
|
Wfdc18
|
WAP four-disulfide core domain 18 |
| chr9_+_61738471 | 3.82 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr7_+_141973553 | 3.81 |
ENSRNOT00000052075
|
Mettl7a
|
methyltransferase like 7A |
| chr7_+_61165640 | 3.76 |
ENSRNOT00000087955
|
Mdm1
|
Mdm1 nuclear protein |
| chr16_+_55834458 | 3.59 |
ENSRNOT00000086141
|
AABR07025928.1
|
|
| chr5_+_159220979 | 3.44 |
ENSRNOT00000008940
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr5_-_57683932 | 3.44 |
ENSRNOT00000074796
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr18_+_70248713 | 3.38 |
ENSRNOT00000032202
|
Mbd1
|
methyl-CpG binding domain protein 1 |
| chr5_-_162602339 | 3.34 |
ENSRNOT00000071779
|
RGD1563334
|
similar to novel protein similar to esterases |
| chr5_-_160158386 | 3.04 |
ENSRNOT00000089345
|
Fblim1
|
filamin binding LIM protein 1 |
| chr5_-_157820889 | 2.98 |
ENSRNOT00000024326
|
Mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr11_+_67451549 | 2.95 |
ENSRNOT00000042777
|
Stfa2l2
|
stefin A2-like 2 |
| chr19_-_25220010 | 2.93 |
ENSRNOT00000008786
|
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr10_-_61045355 | 2.84 |
ENSRNOT00000048167
|
Olr1514
|
olfactory receptor 1514 |
| chr7_+_23854846 | 2.59 |
ENSRNOT00000037290
|
Bpifc
|
BPI fold containing family C |
| chr1_-_90151405 | 2.42 |
ENSRNOT00000028688
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr7_+_58366192 | 2.37 |
ENSRNOT00000081891
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr2_-_34923024 | 2.36 |
ENSRNOT00000090060
|
Cwc27
|
CWC27 spliceosome-associated protein homolog |
| chr9_+_20004280 | 2.33 |
ENSRNOT00000075670
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr1_-_80271001 | 2.31 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr2_-_34419428 | 2.29 |
ENSRNOT00000048123
|
AABR07007905.1
|
|
| chr3_-_111587151 | 2.15 |
ENSRNOT00000007299
|
Rpap1
|
RNA polymerase II associated protein 1 |
| chr5_-_135859643 | 2.14 |
ENSRNOT00000024719
|
Urod
|
uroporphyrinogen decarboxylase |
| chr5_+_68717519 | 2.12 |
ENSRNOT00000066226
|
Smc2
|
structural maintenance of chromosomes 2 |
| chr13_-_78851600 | 2.09 |
ENSRNOT00000076985
|
Zbtb37
|
zinc finger and BTB domain containing 37 |
| chr4_+_106361762 | 1.92 |
ENSRNOT00000052417
|
AABR07061136.1
|
|
| chr2_+_187512164 | 1.84 |
ENSRNOT00000051394
|
Mef2d
|
myocyte enhancer factor 2D |
| chr14_+_17123360 | 1.71 |
ENSRNOT00000003070
|
Nup54
|
nucleoporin 54 |
| chrX_-_111102464 | 1.57 |
ENSRNOT00000084176
|
Ripply1
|
ripply transcriptional repressor 1 |
| chr12_-_16310190 | 1.53 |
ENSRNOT00000060444
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr4_-_167180656 | 1.46 |
ENSRNOT00000049487
|
Tas2r103
|
taste receptor, type 2, member 103 |
| chr1_+_163797660 | 1.18 |
ENSRNOT00000021674
|
Wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr4_-_69149520 | 1.02 |
ENSRNOT00000017611
|
Prss58
|
protease, serine, 58 |
| chr7_-_14369821 | 0.97 |
ENSRNOT00000008560
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chr9_+_14951047 | 0.87 |
ENSRNOT00000074267
|
LOC100911489
|
uncharacterized LOC100911489 |
| chr2_+_257295193 | 0.79 |
ENSRNOT00000052249
|
AABR07013798.1
|
|
| chr3_+_75809445 | 0.70 |
ENSRNOT00000049950
|
Olr581
|
olfactory receptor 581 |
| chr2_+_192553983 | 0.69 |
ENSRNOT00000085204
|
LOC102550416
|
small proline-rich protein 2I-like |
| chr5_+_157820908 | 0.55 |
ENSRNOT00000068168
|
Emc1
|
ER membrane protein complex subunit 1 |
| chr4_+_152437966 | 0.49 |
ENSRNOT00000013070
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr7_-_143085833 | 0.48 |
ENSRNOT00000031257
ENSRNOT00000077286 |
Krt83
|
keratin 83 |
| chr17_-_71897972 | 0.44 |
ENSRNOT00000065942
|
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr1_+_63201271 | 0.42 |
ENSRNOT00000090830
|
LOC108348164
|
vomeronasal type-1 receptor 4-like |
| chr4_+_1535100 | 0.23 |
ENSRNOT00000074575
|
LOC100912488
|
olfactory receptor 147-like |
| chr9_-_44617210 | 0.23 |
ENSRNOT00000074343
|
Lyg1
|
lysozyme G1 |
| chr3_+_147073160 | 0.15 |
ENSRNOT00000012690
|
Sdcbp2
|
syndecan binding protein 2 |
| chr1_-_168869420 | 0.05 |
ENSRNOT00000021436
|
Olr125
|
olfactory receptor 125 |
| chr1_+_207508414 | 0.01 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mga
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 3.6 | 17.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 3.5 | 10.6 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) |
| 2.9 | 14.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 2.4 | 7.3 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 2.2 | 6.5 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 2.1 | 6.4 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 2.1 | 10.3 | GO:0036337 | ovarian follicle atresia(GO:0001552) Fas signaling pathway(GO:0036337) |
| 2.0 | 8.1 | GO:2001038 | senescence-associated heterochromatin focus assembly(GO:0035986) histone H2A phosphorylation(GO:1990164) regulation of cellular response to drug(GO:2001038) |
| 1.9 | 13.0 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) oxidative stress-induced premature senescence(GO:0090403) |
| 1.8 | 16.6 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 1.8 | 20.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.6 | 9.6 | GO:0048597 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 1.5 | 5.9 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 1.1 | 10.8 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 1.1 | 4.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 1.0 | 6.1 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) |
| 1.0 | 10.0 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 1.0 | 25.2 | GO:0030220 | platelet formation(GO:0030220) |
| 0.7 | 7.7 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.7 | 32.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.7 | 10.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.6 | 5.7 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.6 | 15.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.6 | 1.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.6 | 3.4 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.6 | 5.0 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.5 | 12.4 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.5 | 3.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.5 | 2.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 4.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.4 | 8.7 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 10.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 15.4 | GO:0007140 | male meiosis(GO:0007140) |
| 0.3 | 1.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.3 | 9.1 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.2 | 3.0 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 2.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.2 | 1.7 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 3.4 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.2 | 6.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 1.0 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 6.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 8.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 3.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.8 | GO:0007512 | adult heart development(GO:0007512) |
| 0.1 | 6.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 3.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 4.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 15.0 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 8.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 9.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 2.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.5 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 21.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 2.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 7.6 | GO:0032886 | regulation of microtubule-based process(GO:0032886) |
| 0.1 | 3.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 5.6 | GO:0007369 | gastrulation(GO:0007369) |
| 0.0 | 1.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 4.5 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0005712 | chiasma(GO:0005712) recombination nodule(GO:0005713) |
| 2.1 | 10.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 1.8 | 12.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 1.6 | 8.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.3 | 6.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.3 | 3.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.8 | 5.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.5 | 9.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 6.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 9.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 14.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 2.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 6.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 3.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 10.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 11.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 2.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 1.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 4.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 10.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 4.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 3.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 3.0 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 4.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 13.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 7.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 8.9 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 16.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 22.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 4.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 9.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 3.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 13.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 3.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 6.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 7.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 25.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 52.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 6.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 9.3 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 2.5 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 17.9 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 3.6 | 14.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 2.1 | 10.6 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 2.1 | 10.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 2.0 | 10.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 2.0 | 6.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 2.0 | 8.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 2.0 | 5.9 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) leukotriene-C4 synthase activity(GO:0004464) |
| 1.9 | 7.7 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 1.5 | 15.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 1.2 | 7.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.1 | 9.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.1 | 6.5 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.0 | 12.4 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.8 | 6.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 9.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.6 | 15.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.6 | 13.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.6 | 3.4 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.5 | 14.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 3.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 6.7 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.3 | 3.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 25.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 19.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.2 | 10.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 4.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 15.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 9.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 6.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 10.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 3.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 1.0 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 25.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 15.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 2.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 7.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.8 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 4.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 6.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 9.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.9 | 10.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.7 | 17.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.7 | 9.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.6 | 6.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.5 | 32.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.3 | 11.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 12.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 29.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 10.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 6.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 6.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 20.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.9 | 14.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.8 | 14.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.7 | 10.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.6 | 17.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.5 | 6.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 6.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 8.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 9.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 12.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 10.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 3.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 7.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 14.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |