Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
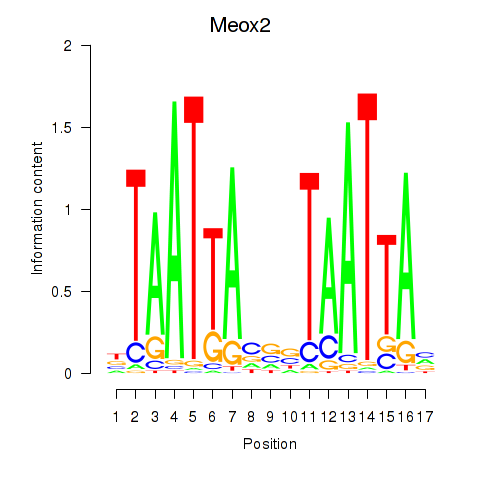
Results for Meox2
Z-value: 1.38
Transcription factors associated with Meox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox2
|
ENSRNOG00000006588 | mesenchyme homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox2 | rn6_v1_chr6_+_56625650_56625650 | 0.45 | 1.1e-17 | Click! |
Activity profile of Meox2 motif
Sorted Z-values of Meox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrM_+_10160 | 40.99 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr4_-_129619142 | 40.86 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chrM_+_9870 | 39.03 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr2_+_248398917 | 31.70 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr5_+_6373583 | 31.69 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr10_+_53713938 | 31.51 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr6_-_143065639 | 28.27 |
ENSRNOT00000070923
|
AABR07065827.1
|
|
| chrM_+_9451 | 27.52 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr3_+_16846412 | 25.85 |
ENSRNOT00000074266
|
AABR07051551.1
|
|
| chrX_-_64715823 | 22.47 |
ENSRNOT00000076297
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr2_-_211017778 | 21.92 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chrM_+_11736 | 21.83 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr7_-_107392972 | 21.82 |
ENSRNOT00000093425
|
Tmem71
|
transmembrane protein 71 |
| chr4_+_51614676 | 21.31 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_+_116632506 | 21.16 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr9_-_73871888 | 20.89 |
ENSRNOT00000017686
|
Acadl
|
acyl-CoA dehydrogenase, long chain |
| chr2_+_42829413 | 19.51 |
ENSRNOT00000065528
|
Actbl2
|
actin, beta-like 2 |
| chr10_+_31880918 | 19.11 |
ENSRNOT00000059448
|
Timd4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr4_+_166833320 | 19.09 |
ENSRNOT00000031438
|
Tas2r120
|
taste receptor, type 2, member 120 |
| chr17_+_32973695 | 18.27 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr2_-_261337163 | 18.25 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chr4_-_163890801 | 17.58 |
ENSRNOT00000081946
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr2_-_203680083 | 16.82 |
ENSRNOT00000021268
|
Cd2
|
Cd2 molecule |
| chr19_-_49448072 | 16.30 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr13_-_113927745 | 15.85 |
ENSRNOT00000010910
ENSRNOT00000080616 |
Cr2
|
complement C3d receptor 2 |
| chr6_+_76349362 | 14.85 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr8_+_100260049 | 14.70 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr9_-_97151832 | 14.65 |
ENSRNOT00000040169
|
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr11_-_45124423 | 13.85 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr4_+_98481520 | 13.58 |
ENSRNOT00000078381
ENSRNOT00000048493 |
AABR07060886.1
|
|
| chr4_+_109467272 | 13.49 |
ENSRNOT00000008212
|
Reg3b
|
regenerating family member 3 beta |
| chr11_-_75144903 | 13.31 |
ENSRNOT00000002329
|
Hrasls
|
HRAS-like suppressor |
| chr20_-_45260119 | 13.15 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr4_+_162493908 | 13.10 |
ENSRNOT00000072064
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr3_+_16571602 | 12.92 |
ENSRNOT00000048351
|
LOC100361705
|
rCG64259-like |
| chr6_-_21600451 | 12.90 |
ENSRNOT00000047674
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_16441030 | 12.70 |
ENSRNOT00000047784
|
AABR07051532.1
|
|
| chr7_+_117409576 | 12.62 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr4_-_163762434 | 12.43 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chrX_+_78372257 | 12.06 |
ENSRNOT00000046164
|
Gpr174
|
G protein-coupled receptor 174 |
| chr1_-_77865870 | 12.05 |
ENSRNOT00000017353
|
Ehd2
|
EH-domain containing 2 |
| chr17_-_31780120 | 11.64 |
ENSRNOT00000058388
|
AABR07027450.1
|
|
| chr6_-_141321108 | 11.25 |
ENSRNOT00000040556
|
AABR07065789.3
|
|
| chr15_+_55461695 | 11.23 |
ENSRNOT00000065195
|
Sucla2
|
succinate-CoA ligase ADP-forming beta subunit |
| chr15_-_29548400 | 11.17 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr4_-_103024841 | 11.09 |
ENSRNOT00000090283
|
AABR07061036.1
|
|
| chr11_+_85430400 | 11.08 |
ENSRNOT00000083198
|
AABR07034729.1
|
|
| chr17_-_15519060 | 11.03 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr6_-_139997537 | 10.89 |
ENSRNOT00000073207
|
AABR07065740.1
|
|
| chrM_+_8599 | 10.67 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr3_-_16999720 | 10.62 |
ENSRNOT00000074382
|
RGD1563231
|
similar to immunoglobulin kappa-chain VK-1 |
| chr10_-_98388637 | 10.59 |
ENSRNOT00000005665
|
Abca8a
|
ATP-binding cassette, subfamily A (ABC1), member 8a |
| chr3_+_16753703 | 10.23 |
ENSRNOT00000077741
|
AABR07051548.2
|
|
| chr13_-_83202864 | 10.06 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr6_-_138250089 | 9.80 |
ENSRNOT00000048378
|
Ighm
|
immunoglobulin heavy constant mu |
| chr4_+_64088900 | 9.75 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr3_+_17139670 | 9.47 |
ENSRNOT00000073316
|
AABR07051564.1
|
|
| chr2_+_41442241 | 9.40 |
ENSRNOT00000067546
|
Pde4d
|
phosphodiesterase 4D |
| chr9_-_50762082 | 9.32 |
ENSRNOT00000015492
|
Mettl21c
|
methyltransferase like 21C |
| chrX_+_77065397 | 9.13 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr15_-_34693034 | 9.03 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr7_-_29233392 | 8.88 |
ENSRNOT00000064241
|
Spic
|
Spi-C transcription factor |
| chr3_+_19772056 | 8.81 |
ENSRNOT00000044455
|
AABR07051708.1
|
|
| chr8_-_87315955 | 8.77 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chr20_+_4020317 | 8.77 |
ENSRNOT00000000526
|
RT1-DOb
|
RT1 class II, locus DOb |
| chr11_-_17684903 | 8.60 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr4_-_159399634 | 8.50 |
ENSRNOT00000089193
|
Ndufa9
|
NADH:ubiquinone oxidoreductase subunit A9 |
| chr6_+_108009251 | 8.44 |
ENSRNOT00000059073
|
Ptgr2
|
prostaglandin reductase 2 |
| chr10_+_30038709 | 8.42 |
ENSRNOT00000005898
|
Il12b
|
interleukin 12B |
| chr1_-_14412807 | 8.40 |
ENSRNOT00000074583
|
Tnfaip3
|
TNF alpha induced protein 3 |
| chr3_+_97349454 | 8.25 |
ENSRNOT00000089524
|
Dcdc5
|
doublecortin domain containing 5 |
| chr4_+_147275334 | 8.25 |
ENSRNOT00000051858
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr11_+_15081774 | 8.16 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chrM_+_3904 | 8.03 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr7_+_38897278 | 8.02 |
ENSRNOT00000006450
|
Epyc
|
epiphycan |
| chr10_-_98469799 | 7.97 |
ENSRNOT00000087502
ENSRNOT00000088646 |
Abca9
|
ATP binding cassette subfamily A member 9 |
| chr6_-_123577695 | 7.95 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chrX_+_134979646 | 7.91 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr4_-_103569159 | 7.89 |
ENSRNOT00000084036
|
AABR07061072.1
|
|
| chr6_-_143702033 | 7.79 |
ENSRNOT00000051410
|
AABR07065886.1
|
|
| chr7_+_118685022 | 7.73 |
ENSRNOT00000089135
|
Apol3
|
apolipoprotein L, 3 |
| chrX_+_158350347 | 7.67 |
ENSRNOT00000066874
|
LOC100909732
|
protein FAM122B-like |
| chr3_+_21016266 | 7.62 |
ENSRNOT00000010473
|
Olr420
|
olfactory receptor 420 |
| chrX_+_31984612 | 7.60 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr5_+_166870011 | 7.59 |
ENSRNOT00000088299
|
Rps2
|
ribosomal protein S2 |
| chr7_-_18793289 | 7.48 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chrX_+_83678339 | 7.37 |
ENSRNOT00000005997
|
Apool
|
apolipoprotein O-like |
| chrM_+_7919 | 7.27 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr7_-_12432130 | 7.13 |
ENSRNOT00000077301
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr4_+_102351036 | 7.12 |
ENSRNOT00000079277
|
AABR07060994.1
|
|
| chr10_-_34242985 | 7.07 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr3_-_98092131 | 7.02 |
ENSRNOT00000006512
|
Fshb
|
follicle stimulating hormone beta subunit |
| chr8_+_42322813 | 7.01 |
ENSRNOT00000040431
|
Olr1241
|
olfactory receptor 1241 |
| chr13_+_51229890 | 6.99 |
ENSRNOT00000060669
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr13_-_91776397 | 6.98 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chrX_+_121612952 | 6.95 |
ENSRNOT00000022122
|
AABR07041179.1
|
|
| chr4_-_167202106 | 6.79 |
ENSRNOT00000038581
|
Tas2r140
|
taste receptor, type 2, member 140 |
| chr6_-_142676432 | 6.66 |
ENSRNOT00000074947
|
AABR07065815.2
|
|
| chr3_-_74545241 | 6.60 |
ENSRNOT00000090836
|
Olr531
|
olfactory receptor 531 |
| chr1_-_90149991 | 6.53 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr3_+_44025300 | 6.51 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chrM_+_7758 | 6.51 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr18_-_74059533 | 6.50 |
ENSRNOT00000038767
|
RGD1308601
|
similar to hypothetical protein |
| chr13_-_37697569 | 6.46 |
ENSRNOT00000045263
|
LOC680288
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr4_-_167180656 | 6.43 |
ENSRNOT00000049487
|
Tas2r103
|
taste receptor, type 2, member 103 |
| chr13_+_92146586 | 6.34 |
ENSRNOT00000004659
|
Olr1588
|
olfactory receptor 1588 |
| chr8_-_109621408 | 6.30 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr14_+_108826831 | 6.26 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr17_+_36451058 | 6.19 |
ENSRNOT00000071408
|
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr17_-_43640387 | 6.18 |
ENSRNOT00000087731
|
Hist1h1c
|
histone cluster 1 H1 family member c |
| chr6_+_122254841 | 6.14 |
ENSRNOT00000005055
|
Gpr65
|
G-protein coupled receptor 65 |
| chr6_-_139102378 | 6.13 |
ENSRNOT00000086423
|
AABR07065656.5
|
|
| chr15_-_35113678 | 6.10 |
ENSRNOT00000059677
|
Ctsg
|
cathepsin G |
| chr12_-_19439977 | 6.08 |
ENSRNOT00000060035
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr7_-_3707226 | 6.05 |
ENSRNOT00000064380
|
Olr879
|
olfactory receptor 879 |
| chr1_-_173180745 | 6.04 |
ENSRNOT00000043904
|
Olr203
|
olfactory receptor 203 |
| chr1_+_228142778 | 6.02 |
ENSRNOT00000028517
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr5_+_16845631 | 5.97 |
ENSRNOT00000047889
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_+_11763706 | 5.89 |
ENSRNOT00000078366
|
Sh2d3c
|
SH2 domain containing 3C |
| chr1_-_242152834 | 5.86 |
ENSRNOT00000071614
ENSRNOT00000020412 |
Fxn
|
frataxin |
| chr3_-_94808861 | 5.85 |
ENSRNOT00000038464
|
Prrg4
|
proline rich and Gla domain 4 |
| chr17_-_31916553 | 5.79 |
ENSRNOT00000074220
|
LOC100911032
|
uncharacterized LOC100911032 |
| chr3_+_55461420 | 5.77 |
ENSRNOT00000073549
|
G6pc2
|
glucose-6-phosphatase catalytic subunit 2 |
| chr17_-_54808483 | 5.71 |
ENSRNOT00000075683
|
AABR07028049.1
|
|
| chr4_+_102290338 | 5.71 |
ENSRNOT00000011067
|
AABR07060992.1
|
|
| chr10_+_60496864 | 5.60 |
ENSRNOT00000081152
|
Olfr390
|
olfactory receptor 390 |
| chr15_+_31948035 | 5.60 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr11_-_32088002 | 5.58 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr2_-_14701903 | 5.57 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr4_-_164051812 | 5.51 |
ENSRNOT00000085719
|
AABR07062183.1
|
|
| chr11_-_62067655 | 5.46 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_-_72805848 | 5.44 |
ENSRNOT00000009040
|
Gpr33
|
G protein-coupled receptor 33 |
| chr4_-_157154120 | 5.41 |
ENSRNOT00000056058
|
C1s
|
complement C1s |
| chr13_+_91954138 | 5.35 |
ENSRNOT00000074608
ENSRNOT00000037655 |
Aim2
|
absent in melanoma 2 |
| chr4_-_170912629 | 5.26 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr17_-_70548071 | 5.26 |
ENSRNOT00000073144
|
Il2ra
|
interleukin 2 receptor subunit alpha |
| chr6_-_143206772 | 5.24 |
ENSRNOT00000073713
|
AABR07065839.1
|
|
| chr3_+_18970574 | 5.24 |
ENSRNOT00000088394
|
AABR07051665.1
|
|
| chr3_-_73510015 | 5.18 |
ENSRNOT00000072248
|
Olr483
|
olfactory receptor 483 |
| chr4_+_1459363 | 5.15 |
ENSRNOT00000082919
|
Olr1234
|
olfactory receptor 1234 |
| chr2_-_170301348 | 5.07 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr7_-_74735650 | 5.07 |
ENSRNOT00000014407
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr16_+_54358471 | 5.05 |
ENSRNOT00000047314
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr19_+_3325893 | 5.02 |
ENSRNOT00000048879
|
RGD1565617
|
similar to Ig variable region, light chain |
| chr4_-_69211671 | 5.01 |
ENSRNOT00000017852
|
LOC286960
|
preprotrypsinogen IV |
| chr10_-_87195075 | 5.00 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chr15_+_30621374 | 4.98 |
ENSRNOT00000090632
|
AABR07017748.2
|
|
| chr4_-_164174725 | 4.97 |
ENSRNOT00000080754
|
AABR07062185.1
|
|
| chr11_+_76833761 | 4.97 |
ENSRNOT00000042650
|
Uts2b
|
urotensin 2B |
| chr1_-_211265161 | 4.93 |
ENSRNOT00000080041
ENSRNOT00000023477 |
Bnip3
|
BCL2 interacting protein 3 |
| chr4_+_1671269 | 4.88 |
ENSRNOT00000071452
|
Olr1251
|
olfactory receptor 1251 |
| chr5_+_121952977 | 4.87 |
ENSRNOT00000008278
|
Pde4b
|
phosphodiesterase 4B |
| chr15_-_18521161 | 4.85 |
ENSRNOT00000051556
|
LOC498460
|
LRRGT00055 |
| chr6_-_142299996 | 4.79 |
ENSRNOT00000086580
|
Ighv8-2
|
immunoglobulin heavy variable V8-2 |
| chr15_-_12262164 | 4.79 |
ENSRNOT00000008798
|
Olr1606
|
olfactory receptor 1606 |
| chr8_+_117117430 | 4.77 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr3_+_17180411 | 4.73 |
ENSRNOT00000058260
|
AABR07051565.1
|
|
| chr17_-_81002838 | 4.69 |
ENSRNOT00000089807
|
St8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr15_+_29096865 | 4.68 |
ENSRNOT00000077709
|
AABR07017597.2
|
|
| chr3_-_74597393 | 4.68 |
ENSRNOT00000013201
|
Olr536
|
olfactory receptor 536 |
| chr11_-_43022565 | 4.60 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr15_-_36302945 | 4.60 |
ENSRNOT00000075675
|
LOC100910599
|
olfactory receptor 144-like |
| chr2_+_233909142 | 4.59 |
ENSRNOT00000073197
|
Dcm5
|
Dcm5 protein |
| chr9_+_98279022 | 4.56 |
ENSRNOT00000038536
|
Rbm44
|
RNA binding motif protein 44 |
| chr1_-_171060011 | 4.48 |
ENSRNOT00000042829
|
Olr226
|
olfactory receptor 226 |
| chr1_+_63333283 | 4.48 |
ENSRNOT00000086294
|
Vom1r30
|
vomeronasal 1 receptor 30 |
| chr8_+_133029625 | 4.41 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr10_-_87521514 | 4.39 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr3_+_103248396 | 4.37 |
ENSRNOT00000043710
|
LOC100909613
|
olfactory receptor 4F15-like |
| chr1_-_172395872 | 4.34 |
ENSRNOT00000055174
|
Olr252
|
olfactory receptor 252 |
| chr3_-_158920794 | 4.30 |
ENSRNOT00000074921
|
Olr1252
|
olfactory receptor 1252 |
| chr8_-_19180021 | 4.28 |
ENSRNOT00000035560
|
LOC100911988
|
olfactory receptor 7G1-like |
| chr15_+_31469953 | 4.27 |
ENSRNOT00000077849
|
AABR07017825.4
|
|
| chr6_-_142880382 | 4.24 |
ENSRNOT00000074706
ENSRNOT00000072861 |
AABR07065821.1
|
|
| chr1_-_227932603 | 4.24 |
ENSRNOT00000033795
|
Ms4a6a
|
membrane spanning 4-domains A6A |
| chr4_+_61924013 | 4.23 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr3_+_75390030 | 4.21 |
ENSRNOT00000037577
|
Olr559
|
olfactory receptor 559 |
| chr18_-_31749647 | 4.20 |
ENSRNOT00000044287
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr9_-_23493081 | 4.12 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr2_+_195684210 | 4.11 |
ENSRNOT00000028316
|
mrpl9
|
mitochondrial ribosomal protein L9 |
| chr4_+_148476943 | 4.10 |
ENSRNOT00000017689
|
Olr823
|
olfactory receptor 823 |
| chr3_-_74607059 | 4.07 |
ENSRNOT00000047002
|
Olr537
|
olfactory receptor 537 |
| chr1_-_149770781 | 4.05 |
ENSRNOT00000082185
|
Olr16
|
olfactory receptor 16 |
| chr4_+_106289816 | 4.05 |
ENSRNOT00000040075
|
AABR07061131.1
|
|
| chr4_+_1658278 | 4.01 |
ENSRNOT00000073845
|
Olr1250
|
olfactory receptor 1250 |
| chr4_+_87943152 | 3.96 |
ENSRNOT00000080536
|
Vom1r78
|
vomeronasal 1 receptor 78 |
| chr14_+_82769642 | 3.96 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr10_-_87564327 | 3.96 |
ENSRNOT00000064760
ENSRNOT00000068237 |
LOC680160
|
similar to keratin associated protein 4-7 |
| chr3_-_76187045 | 3.91 |
ENSRNOT00000075650
|
LOC686683
|
similar to olfactory receptor 73 |
| chr14_-_44375804 | 3.91 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr1_+_213636093 | 3.91 |
ENSRNOT00000019642
|
Psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr10_-_102200596 | 3.90 |
ENSRNOT00000081519
|
Fam104a
|
family with sequence similarity 104, member A |
| chr14_+_62609484 | 3.87 |
ENSRNOT00000003017
|
Guf1
|
GUF1 homolog, GTPase |
| chr2_+_46066780 | 3.86 |
ENSRNOT00000073226
|
LOC108350225
|
olfactory receptor 8B8-like |
| chr6_+_138432550 | 3.84 |
ENSRNOT00000056829
|
Adam6
|
a disintegrin and metallopeptidase domain 6 |
| chr3_-_103265059 | 3.84 |
ENSRNOT00000073965
|
LOC103691873
|
putative olfactory receptor GPCRLTM7 |
| chr4_+_1363724 | 3.83 |
ENSRNOT00000072782
|
LOC100910863
|
olfactory receptor 143-like |
| chr3_+_19366370 | 3.81 |
ENSRNOT00000086557
|
AABR07051689.1
|
|
| chr3_+_77047466 | 3.78 |
ENSRNOT00000089424
|
Olr648
|
olfactory receptor 648 |
| chr1_+_229849892 | 3.77 |
ENSRNOT00000072886
|
Olr346
|
olfactory receptor 346 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 7.0 | 20.9 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 5.3 | 31.7 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 5.1 | 40.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 4.6 | 18.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 4.2 | 8.4 | GO:0070425 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 4.0 | 12.0 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 3.4 | 13.5 | GO:1903576 | response to L-arginine(GO:1903576) |
| 3.4 | 10.1 | GO:0071663 | immunoglobulin production in mucosal tissue(GO:0002426) T-helper 1 cell activation(GO:0035711) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) regulation of thymocyte migration(GO:2000410) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 3.1 | 9.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 2.9 | 8.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 2.8 | 11.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 2.7 | 8.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 2.6 | 13.2 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 2.4 | 31.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 2.1 | 49.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 2.1 | 8.4 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 2.1 | 6.3 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 2.1 | 6.3 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 2.1 | 6.2 | GO:0035600 | tRNA methylthiolation(GO:0035600) |
| 2.0 | 6.1 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 2.0 | 5.9 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) proprioception(GO:0019230) |
| 2.0 | 9.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.9 | 28.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 1.7 | 16.8 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 1.6 | 4.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.6 | 4.8 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 1.6 | 62.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 1.3 | 5.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.2 | 4.9 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.2 | 3.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 1.2 | 3.5 | GO:0045399 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.1 | 13.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.1 | 3.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.0 | 10.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 1.0 | 4.0 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.0 | 18.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.9 | 4.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.9 | 6.5 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.9 | 12.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.8 | 20.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.8 | 0.8 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.8 | 6.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.8 | 5.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.7 | 4.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.7 | 7.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.6 | 6.8 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.6 | 7.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.6 | 3.1 | GO:0010958 | regulation of amino acid import(GO:0010958) glycine import(GO:0036233) |
| 0.6 | 7.9 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.6 | 8.5 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.6 | 4.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 1.6 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.5 | 2.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.5 | 2.6 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.5 | 3.6 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.5 | 10.1 | GO:0009750 | response to fructose(GO:0009750) |
| 0.5 | 4.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.5 | 1.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.5 | 39.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.5 | 2.7 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.5 | 2.7 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.4 | 7.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.4 | 21.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.4 | 1.6 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.4 | 3.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 4.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.3 | 8.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 1.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.3 | 3.7 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 3.0 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.3 | 3.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.3 | 4.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.3 | 6.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.3 | 4.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.3 | 336.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 12.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 3.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 2.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 8.4 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.2 | 8.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.2 | 0.9 | GO:0052422 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.2 | 2.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 11.0 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 2.8 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.2 | 2.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 3.4 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.2 | 2.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 0.8 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.2 | 5.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 5.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 1.0 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 1.9 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.2 | 1.9 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 1.1 | GO:0046687 | response to chromate(GO:0046687) |
| 0.1 | 1.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 11.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 10.3 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 7.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 6.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.9 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 4.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 5.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 4.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 1.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 1.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 3.1 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 22.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.1 | 0.8 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 1.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.1 | 1.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 | 45.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.1 | 2.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 3.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 7.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 3.7 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 1.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.7 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.0 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 2.2 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.5 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 1.6 | GO:1903955 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 1.9 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 3.4 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.8 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 0.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 1.1 | GO:0008585 | female gonad development(GO:0008585) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 31.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 3.5 | 31.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 3.0 | 152.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.6 | 9.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.3 | 13.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.3 | 19.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 1.2 | 21.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 1.2 | 12.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 1.2 | 19.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 1.1 | 5.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 1.1 | 7.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.0 | 4.8 | GO:0097413 | Lewy body(GO:0097413) |
| 0.9 | 21.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.9 | 7.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.8 | 8.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.6 | 33.3 | GO:0031672 | A band(GO:0031672) |
| 0.6 | 1.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.6 | 16.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.5 | 8.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 13.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.4 | 3.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 1.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 4.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 6.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 12.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 12.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 2.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 4.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 6.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 6.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 3.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 8.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 3.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 11.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 9.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 4.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 7.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 7.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.9 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 3.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.9 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 18.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 5.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 6.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.0 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 15.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 4.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 8.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 14.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 36.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 135.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 3.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.6 | GO:0005840 | ribosome(GO:0005840) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 31.7 | GO:0019002 | GMP binding(GO:0019002) |
| 7.7 | 23.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 5.2 | 145.9 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 3.7 | 11.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 3.5 | 21.2 | GO:0035473 | lipase binding(GO:0035473) |
| 3.3 | 9.8 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 3.0 | 18.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 2.8 | 8.4 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 2.7 | 40.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 2.7 | 16.2 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 2.4 | 26.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 2.2 | 6.7 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.1 | 14.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.1 | 6.2 | GO:0050497 | methylthiotransferase activity(GO:0035596) transferase activity, transferring alkylthio groups(GO:0050497) |
| 1.8 | 5.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 1.4 | 8.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.2 | 8.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 1.2 | 7.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.2 | 5.8 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.1 | 1.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 1.1 | 4.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.0 | 5.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.9 | 4.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.9 | 2.7 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.9 | 9.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.8 | 12.1 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.7 | 3.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.7 | 19.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 12.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.6 | 6.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.6 | 3.5 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.6 | 3.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.5 | 1.6 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.5 | 2.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.5 | 4.1 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.5 | 197.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.5 | 2.8 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.5 | 4.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.5 | 6.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 12.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.4 | 4.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.4 | 4.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.4 | 8.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.4 | 12.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.4 | 3.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 1.8 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.3 | 6.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 9.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 7.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.3 | 5.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 16.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 0.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 4.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 4.7 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.2 | 2.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 2.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 6.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 2.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 3.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.9 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.2 | 15.9 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.2 | 2.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 9.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.2 | 140.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 7.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 10.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 13.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 1.6 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 7.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 15.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 20.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 3.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 5.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 11.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 3.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 1.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 14.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 6.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 5.5 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.1 | 4.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 3.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 14.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 6.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 5.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 1.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 3.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 26.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.3 | 8.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 8.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 13.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 13.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.2 | 7.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 5.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 11.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 4.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 9.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 4.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 20.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.1 | 6.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 11.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.8 | 12.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.7 | 7.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 40.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.6 | 6.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 30.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.5 | 5.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 4.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 14.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 8.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 3.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.3 | 14.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 5.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 4.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 8.4 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.2 | 2.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 44.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 12.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 12.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 5.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 6.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 5.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 5.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 6.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 4.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 5.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 4.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 2.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 3.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |