Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Meox1
Z-value: 0.66
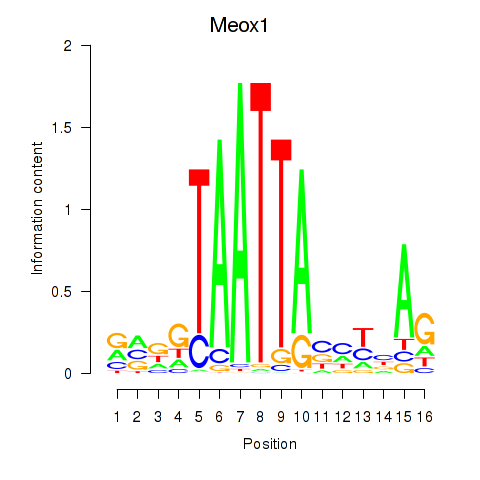
Transcription factors associated with Meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox1
|
ENSRNOG00000020803 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox1 | rn6_v1_chr10_-_89816491_89816491 | -0.12 | 2.6e-02 | Click! |
Activity profile of Meox1 motif
Sorted Z-values of Meox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_95501778 | 29.60 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr5_-_126911520 | 22.06 |
ENSRNOT00000091521
|
Dio1
|
deiodinase, iodothyronine, type I |
| chr17_-_43537293 | 19.01 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr18_-_6781841 | 18.71 |
ENSRNOT00000077606
ENSRNOT00000048109 |
Aqp4
|
aquaporin 4 |
| chr14_+_22517774 | 16.71 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr5_-_133959447 | 14.89 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr8_-_78233430 | 14.44 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr20_+_40769586 | 14.41 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr12_+_47179664 | 13.81 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr14_+_22375955 | 13.42 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr10_-_47666921 | 12.81 |
ENSRNOT00000086579
|
Slc47a1
|
solute carrier family 47 member 1 |
| chr3_+_48096954 | 12.66 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr7_-_76294663 | 11.85 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr4_-_55011415 | 11.59 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr14_-_64476796 | 11.56 |
ENSRNOT00000029104
|
Gba3
|
glucosidase, beta, acid 3 |
| chr7_-_76488216 | 11.41 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr1_-_67065797 | 11.25 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chrX_+_5825711 | 11.07 |
ENSRNOT00000037360
|
Efhc2
|
EF-hand domain containing 2 |
| chr1_+_217345545 | 10.90 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_-_82533689 | 10.52 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr1_+_248428099 | 10.35 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr1_-_189182306 | 9.59 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr17_+_8489266 | 9.46 |
ENSRNOT00000016252
|
Lect2
|
leukocyte cell-derived chemotaxin 2 |
| chr11_+_57207656 | 8.79 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr8_+_33239139 | 8.60 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_+_6644643 | 8.57 |
ENSRNOT00000051670
|
Olr962
|
olfactory receptor 962 |
| chr1_+_87224677 | 8.35 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chrX_-_69218526 | 8.31 |
ENSRNOT00000092321
ENSRNOT00000074071 ENSRNOT00000092571 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr1_+_59156251 | 8.20 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr18_-_24735349 | 8.19 |
ENSRNOT00000036537
|
Gpr17
|
G protein-coupled receptor 17 |
| chr3_+_63510293 | 8.19 |
ENSRNOT00000058093
|
Dfnb59
|
deafness, autosomal recessive 59 |
| chr20_-_10013190 | 8.09 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr1_-_104024682 | 7.79 |
ENSRNOT00000056081
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr2_-_236480502 | 7.48 |
ENSRNOT00000015020
|
Sgms2
|
sphingomyelin synthase 2 |
| chr2_+_69415057 | 7.35 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr1_+_88955135 | 7.22 |
ENSRNOT00000083550
|
Prodh2
|
proline dehydrogenase 2 |
| chr2_+_22950018 | 7.16 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chrX_+_84064427 | 6.94 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr8_-_7426611 | 6.84 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr2_-_40386669 | 6.83 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr1_+_219833299 | 6.83 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr20_-_10013559 | 6.69 |
ENSRNOT00000091623
|
Rsph1
|
radial spoke head 1 homolog |
| chr11_+_44072043 | 6.53 |
ENSRNOT00000033716
|
LOC680520
|
similar to CG4349-PA |
| chrX_+_6273733 | 6.38 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr5_-_173233188 | 6.30 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr6_+_60566196 | 5.93 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chr16_+_74865516 | 5.84 |
ENSRNOT00000058072
|
Atp7b
|
ATPase copper transporting beta |
| chr18_-_67224566 | 5.66 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr17_+_11683862 | 5.39 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr2_+_187162017 | 5.35 |
ENSRNOT00000080693
|
Insrr
|
insulin receptor-related receptor |
| chr7_-_101138373 | 5.35 |
ENSRNOT00000043257
|
LOC500877
|
Ab1-152 |
| chr16_-_10802512 | 5.32 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr9_+_98438439 | 5.15 |
ENSRNOT00000027220
|
Scly
|
selenocysteine lyase |
| chr1_+_48077033 | 5.01 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr2_-_18531210 | 4.97 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr4_-_62860446 | 4.91 |
ENSRNOT00000015752
|
Fam180a
|
family with sequence similarity 180, member A |
| chr7_-_140356209 | 4.86 |
ENSRNOT00000077856
|
Rnd1
|
Rho family GTPase 1 |
| chr12_-_52658275 | 4.85 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr7_-_101138860 | 4.73 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr2_-_158133861 | 4.59 |
ENSRNOT00000090700
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr5_-_161981441 | 4.46 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr5_-_136965191 | 4.30 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr3_+_123106694 | 4.21 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr18_+_17043903 | 4.19 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr4_-_67301102 | 4.19 |
ENSRNOT00000034549
|
Dennd2a
|
DENN domain containing 2A |
| chr4_+_115416580 | 4.18 |
ENSRNOT00000018303
|
Atp6v1b1
|
ATPase H+ transporting V1 subunit B1 |
| chr7_-_52404774 | 4.13 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr5_+_78384444 | 4.01 |
ENSRNOT00000034978
|
LOC500475
|
similar to hypothetical protein 4933430I17 |
| chr7_+_380741 | 3.99 |
ENSRNOT00000091013
|
LOC102554748
|
nidogen-2-like |
| chr12_+_18679789 | 3.99 |
ENSRNOT00000001863
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr9_-_44419998 | 3.86 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr10_+_109665682 | 3.83 |
ENSRNOT00000054963
|
Slc25a10
|
solute carrier family 25 member 10 |
| chr19_-_24614019 | 3.82 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr9_+_84203072 | 3.77 |
ENSRNOT00000018882
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr7_-_92996025 | 3.76 |
ENSRNOT00000076327
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr3_-_26056818 | 3.76 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_+_69841998 | 3.72 |
ENSRNOT00000072231
|
Zfp773-ps1
|
zinc finger protein 773, pseudogene 1 |
| chr1_+_28454966 | 3.63 |
ENSRNOT00000078841
ENSRNOT00000030327 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr6_-_3355339 | 3.63 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr1_+_185863043 | 3.57 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr8_-_39551700 | 3.56 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chrX_+_45965301 | 3.51 |
ENSRNOT00000005141
|
Fam47a
|
family with sequence similarity 47, member A |
| chr18_+_63203063 | 3.48 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr18_+_4084228 | 3.46 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr20_+_3830164 | 3.45 |
ENSRNOT00000045533
ENSRNOT00000084117 |
Col11a2
|
collagen type XI alpha 2 chain |
| chr17_+_87274944 | 3.41 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr7_-_107203897 | 3.41 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr16_+_72010106 | 3.34 |
ENSRNOT00000058330
|
Adam5
|
ADAM metallopeptidase domain 5 |
| chr9_+_99753099 | 3.32 |
ENSRNOT00000052361
|
Olr1350
|
olfactory receptor 1350 |
| chr7_-_36660141 | 3.27 |
ENSRNOT00000012363
|
Nudt4
|
nudix hydrolase 4 |
| chr3_-_57646572 | 3.21 |
ENSRNOT00000076202
|
Mettl8
|
methyltransferase like 8 |
| chr3_-_15278645 | 3.18 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr3_+_148654668 | 3.18 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr18_+_51523758 | 3.15 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr8_+_126975833 | 3.14 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr9_+_73418607 | 3.12 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr20_+_1764794 | 3.11 |
ENSRNOT00000075084
|
Olr1736
|
olfactory receptor 1736 |
| chr17_+_32973695 | 3.11 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr9_-_80167033 | 2.98 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr11_-_29710849 | 2.91 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr6_+_101532518 | 2.78 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr11_+_74834050 | 2.64 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chr15_+_4554603 | 2.62 |
ENSRNOT00000075153
|
Kcnk5
|
potassium two pore domain channel subfamily K member 5 |
| chr11_-_32550539 | 2.56 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr11_-_45667254 | 2.44 |
ENSRNOT00000072466
|
Olr1535
|
olfactory receptor 1535 |
| chr11_-_28979169 | 2.42 |
ENSRNOT00000061601
|
AABR07033580.1
|
|
| chr3_+_95233874 | 2.40 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr2_+_145174876 | 2.39 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr4_-_156563438 | 2.39 |
ENSRNOT00000045305
|
Vom2r51
|
vomeronasal 2 receptor, 51 |
| chr16_-_75107931 | 2.38 |
ENSRNOT00000058066
|
Defb15
|
defensin beta 15 |
| chr13_-_76049363 | 2.38 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr1_+_172822544 | 2.38 |
ENSRNOT00000090018
|
AABR07004960.1
|
|
| chr9_+_99745714 | 2.27 |
ENSRNOT00000022152
|
Olr1349
|
olfactory receptor 1349 |
| chr3_-_160561741 | 2.27 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr2_-_35503638 | 2.23 |
ENSRNOT00000007560
|
LOC100910467
|
olfactory receptor 144-like |
| chr7_-_6971557 | 2.19 |
ENSRNOT00000050153
|
Olr960
|
olfactory receptor 960 |
| chr2_+_66940057 | 2.19 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr20_-_11626876 | 2.10 |
ENSRNOT00000001635
|
LOC100361664
|
keratin associated protein 12-1-like |
| chr8_-_49075892 | 2.10 |
ENSRNOT00000082687
|
Arcn1
|
archain 1 |
| chr10_-_44746549 | 2.10 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chrX_-_84768463 | 2.08 |
ENSRNOT00000088570
|
Chm
|
CHM, Rab escort protein 1 |
| chr2_+_158097843 | 2.06 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr20_+_31234493 | 2.01 |
ENSRNOT00000000675
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr13_+_9479995 | 1.99 |
ENSRNOT00000072979
|
LOC100910887
|
olfactory receptor 6C4-like |
| chrX_+_43831892 | 1.97 |
ENSRNOT00000005120
|
Myl6b
|
myosin light chain 6B |
| chr17_-_42740021 | 1.97 |
ENSRNOT00000023063
|
Prl3a1
|
Prolactin family 3, subfamily a, member 1 |
| chr15_+_28894627 | 1.97 |
ENSRNOT00000017795
|
Olr1643
|
olfactory receptor 1643 |
| chr4_-_167068838 | 1.95 |
ENSRNOT00000029713
|
Tas2r125
|
taste receptor, type 2, member 125 |
| chr2_+_243577082 | 1.84 |
ENSRNOT00000016556
|
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr10_+_91126689 | 1.83 |
ENSRNOT00000004046
|
Nmt1
|
N-myristoyltransferase 1 |
| chr1_+_225037737 | 1.83 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr5_-_102743417 | 1.76 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr11_-_29298339 | 1.76 |
ENSRNOT00000061592
|
AABR07033590.1
|
|
| chr3_+_56056925 | 1.74 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr3_+_75970593 | 1.73 |
ENSRNOT00000075834
|
Olr592
|
olfactory receptor 592 |
| chr4_+_181874861 | 1.72 |
ENSRNOT00000002527
|
Ccdc91
|
coiled-coil domain containing 91 |
| chr15_+_56666012 | 1.71 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr10_+_110445797 | 1.68 |
ENSRNOT00000054920
|
Narf
|
nuclear prelamin A recognition factor |
| chr6_+_124123228 | 1.67 |
ENSRNOT00000005329
|
Psmc1
|
proteasome 26S subunit, ATPase 1 |
| chr2_+_206314213 | 1.64 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr1_-_103811148 | 1.60 |
ENSRNOT00000030162
|
Mrgprx2l
|
MAS-related G protein-coupled receptor, member X2-like |
| chr20_-_29199224 | 1.57 |
ENSRNOT00000071477
|
Mcu
|
mitochondrial calcium uniporter |
| chr8_+_82038967 | 1.54 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr3_-_165700489 | 1.49 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr1_-_170694872 | 1.47 |
ENSRNOT00000075443
|
LOC100911839
|
olfactory receptor 2D3-like |
| chr1_+_230217215 | 1.44 |
ENSRNOT00000072772
|
LOC687088
|
similar to olfactory receptor 1467 |
| chr1_+_229461127 | 1.39 |
ENSRNOT00000035813
|
Olr332
|
olfactory receptor 332 |
| chr3_-_21904133 | 1.38 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr2_-_25235275 | 1.35 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr1_+_168575090 | 1.35 |
ENSRNOT00000048299
|
Olr103
|
olfactory receptor 103 |
| chr1_-_167971151 | 1.34 |
ENSRNOT00000043023
|
Olr53
|
olfactory receptor 53 |
| chr17_-_87826421 | 1.34 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr10_+_88997399 | 1.19 |
ENSRNOT00000027116
|
Mlx
|
MLX, MAX dimerization protein |
| chr15_+_11298478 | 1.17 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr8_+_42663633 | 1.17 |
ENSRNOT00000048537
|
Olr1266
|
olfactory receptor 1266 |
| chrX_+_66314418 | 1.17 |
ENSRNOT00000035405
|
Pgr15l
|
G protein-coupled receptor 15-like |
| chr16_-_12538117 | 1.13 |
ENSRNOT00000046081
|
AABR07024718.1
|
|
| chr3_-_76696107 | 1.12 |
ENSRNOT00000044692
|
Olr629
|
olfactory receptor 629 |
| chr7_+_9294709 | 1.10 |
ENSRNOT00000048396
|
Olr1065
|
olfactory receptor 1065 |
| chr3_-_154627257 | 1.09 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr3_-_73124644 | 1.08 |
ENSRNOT00000012711
|
Olr455
|
olfactory receptor 455 |
| chr2_+_178117466 | 1.05 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr10_+_104582955 | 1.03 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr7_-_145062956 | 1.02 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr1_+_169590308 | 0.98 |
ENSRNOT00000023147
|
Olr155
|
olfactory receptor 155 |
| chr20_-_3751994 | 0.97 |
ENSRNOT00000072288
|
Pou5f1
|
POU class 5 homeobox 1 |
| chr13_+_67545430 | 0.96 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr1_+_86533127 | 0.95 |
ENSRNOT00000052208
|
Vom1r4
|
vomeronasal 1 receptor 4 |
| chr1_-_22488131 | 0.95 |
ENSRNOT00000051416
|
Taar7d
|
trace-amine-associated receptor 7d |
| chr16_+_23553647 | 0.92 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_+_46203776 | 0.92 |
ENSRNOT00000074056
|
LOC108350228
|
olfactory receptor 147-like |
| chr16_+_12510827 | 0.90 |
ENSRNOT00000077763
|
AABR07024716.1
|
|
| chr3_+_103556674 | 0.90 |
ENSRNOT00000050168
|
Olr792
|
olfactory receptor 792 |
| chr3_+_77431389 | 0.88 |
ENSRNOT00000042848
|
Olr660
|
olfactory receptor 660 |
| chr9_+_17920677 | 0.88 |
ENSRNOT00000045763
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr4_+_87312766 | 0.87 |
ENSRNOT00000052126
|
Vom1r71
|
vomeronasal 1 receptor 71 |
| chr4_-_82141385 | 0.87 |
ENSRNOT00000008447
|
Hoxa3
|
homeobox A3 |
| chr3_+_147952879 | 0.85 |
ENSRNOT00000031922
|
Defb20
|
defensin beta 20 |
| chr19_+_9668186 | 0.84 |
ENSRNOT00000016563
|
Cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr1_-_169770188 | 0.81 |
ENSRNOT00000072602
|
Olr165
|
olfactory receptor 165 |
| chr1_+_140998240 | 0.79 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr11_+_70056624 | 0.78 |
ENSRNOT00000002447
|
AC133403.1
|
|
| chr9_+_50966766 | 0.78 |
ENSRNOT00000076636
|
Ercc5
|
ERCC excision repair 5, endonuclease |
| chr7_-_59882077 | 0.76 |
ENSRNOT00000068774
|
Myrfl
|
myelin regulatory factor-like |
| chr1_+_75326217 | 0.76 |
ENSRNOT00000044964
|
Vom1r62
|
vomeronasal 1 receptor 62 |
| chr4_+_72637035 | 0.76 |
ENSRNOT00000007235
|
Olr819
|
olfactory receptor 819 |
| chr1_-_71909138 | 0.71 |
ENSRNOT00000029591
|
Nlrp9
|
NLR family, pyrin domain containing 9 |
| chr2_-_60657712 | 0.71 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr4_+_72502935 | 0.70 |
ENSRNOT00000007168
|
Olr813
|
olfactory receptor 813 |
| chr13_-_89545182 | 0.70 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr3_-_10144388 | 0.69 |
ENSRNOT00000042495
|
Abl1
|
ABL proto-oncogene 1, non-receptor tyrosine kinase |
| chr14_+_22091777 | 0.69 |
ENSRNOT00000072483
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr7_-_6691679 | 0.69 |
ENSRNOT00000075430
|
Olr959
|
olfactory receptor 959 |
| chr16_-_12194118 | 0.65 |
ENSRNOT00000071517
|
RGD1559804
|
similar to hypothetical protein 4930474N05 |
| chr1_+_88113445 | 0.60 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr7_+_2504695 | 0.60 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr7_+_9262982 | 0.59 |
ENSRNOT00000047647
|
Olr1063
|
olfactory receptor 1063 |
| chr4_+_87516218 | 0.57 |
ENSRNOT00000090182
|
Vom1r66
|
vomeronasal 1 receptor 66 |
| chr8_-_87419564 | 0.57 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr11_-_82893845 | 0.56 |
ENSRNOT00000075306
|
LOC100912571
|
eukaryotic translation initiation factor 4 gamma 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.7 | GO:0060354 | negative regulation of cell adhesion molecule production(GO:0060354) |
| 4.3 | 12.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 3.4 | 13.6 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 2.4 | 7.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.4 | 9.6 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 1.9 | 5.8 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 1.9 | 7.6 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.8 | 5.4 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 1.8 | 5.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 1.8 | 5.3 | GO:0048377 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of cardiac ventricle development(GO:1904412) |
| 1.7 | 19.0 | GO:0015747 | urate transport(GO:0015747) |
| 1.7 | 6.8 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 1.4 | 5.7 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 1.4 | 4.2 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 1.4 | 6.8 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.3 | 10.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.3 | 3.8 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 1.2 | 13.8 | GO:0010649 | regulation of cell communication by electrical coupling(GO:0010649) |
| 1.1 | 18.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 1.1 | 7.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.1 | 6.4 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 1.0 | 4.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 1.0 | 4.0 | GO:0002933 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.9 | 2.8 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.8 | 10.9 | GO:0099562 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.8 | 3.3 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.8 | 14.4 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.7 | 3.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.7 | 5.2 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.7 | 2.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.7 | 12.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.6 | 1.8 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.6 | 3.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.6 | 3.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 1.7 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.6 | 4.5 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.5 | 6.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 1.4 | GO:0043311 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.4 | 8.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.4 | 1.5 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.4 | 3.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 9.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.4 | 3.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 3.8 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.4 | 1.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.4 | 5.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.3 | 4.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 1.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.3 | 1.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.3 | 3.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 3.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.3 | 21.1 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.3 | 4.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 29.0 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.2 | 2.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 0.7 | GO:1903210 | transitional stage B cell differentiation(GO:0002332) transitional one stage B cell differentiation(GO:0002333) glomerular visceral epithelial cell apoptotic process(GO:1903210) positive regulation of actin filament binding(GO:1904531) positive regulation of actin binding(GO:1904618) activation of protein kinase C activity(GO:1990051) |
| 0.2 | 3.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 23.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.2 | 0.9 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 2.6 | GO:0051151 | locomotion involved in locomotory behavior(GO:0031987) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 6.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 4.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 0.8 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 2.3 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 | 1.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 6.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 5.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 3.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 8.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 1.8 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 9.5 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 10.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 4.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 2.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.9 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 31.6 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 7.7 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 2.7 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.9 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 2.1 | 12.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.1 | 3.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 1.0 | 14.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.7 | 5.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 10.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 3.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 2.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 1.5 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.5 | 3.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.4 | 16.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.3 | 4.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 4.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 5.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 3.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 11.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 4.2 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.2 | 7.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.7 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 86.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 11.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 9.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.6 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 14.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 5.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 8.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 5.3 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 13.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 2.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 21.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 4.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 13.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 4.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.8 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0017042 | glycosylceramidase activity(GO:0017042) |
| 2.9 | 11.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 2.8 | 22.1 | GO:0008430 | selenium binding(GO:0008430) |
| 2.4 | 7.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 2.3 | 6.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 2.1 | 10.3 | GO:0005534 | galactose binding(GO:0005534) |
| 1.9 | 19.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) efflux transmembrane transporter activity(GO:0015562) salt transmembrane transporter activity(GO:1901702) |
| 1.7 | 23.2 | GO:0015250 | water channel activity(GO:0015250) |
| 1.6 | 21.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.5 | 7.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.3 | 3.8 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 1.3 | 3.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 1.2 | 5.8 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.2 | 3.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 1.1 | 5.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.1 | 4.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.9 | 29.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.9 | 5.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.8 | 10.9 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.8 | 3.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.8 | 23.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.7 | 5.0 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.7 | 2.8 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.6 | 2.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.6 | 7.3 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.6 | 3.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 10.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 1.7 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.5 | 1.6 | GO:0015292 | uniporter activity(GO:0015292) |
| 0.5 | 4.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 18.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 5.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 5.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.4 | 3.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.4 | 12.7 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.3 | 1.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 5.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 6.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 2.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 14.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 8.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 1.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 6.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 3.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 4.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 14.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 5.9 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 3.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 1.0 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 2.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.8 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 1.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 9.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.5 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 4.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 8.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 10.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.0 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 7.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 7.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.8 | GO:0016410 | N-acyltransferase activity(GO:0016410) |
| 0.0 | 2.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 9.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 4.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 12.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.3 | GO:0003924 | GTPase activity(GO:0003924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 9.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 8.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.9 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 18.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.5 | 23.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.2 | 18.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.9 | 10.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.9 | 12.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 8.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.5 | 11.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 5.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 5.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 9.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 11.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 11.2 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.3 | 4.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 4.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 10.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 4.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 5.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 2.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 4.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 5.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 6.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 10.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 5.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |