Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
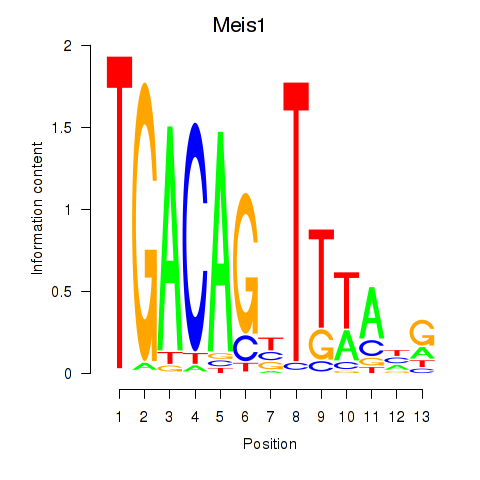
Results for Meis1
Z-value: 1.18
Transcription factors associated with Meis1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meis1
|
ENSRNOG00000004606 | Meis homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis1 | rn6_v1_chr14_-_103321270_103321270 | -0.19 | 6.6e-04 | Click! |
Activity profile of Meis1 motif
Sorted Z-values of Meis1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_80862963 | 67.58 |
ENSRNOT00000004864
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr20_-_45024315 | 60.71 |
ENSRNOT00000066856
|
RGD1304770
|
similar to Na+ dependent glucose transporter 1 |
| chr9_+_4817854 | 47.08 |
ENSRNOT00000040879
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr8_-_49109981 | 46.51 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr5_+_33784715 | 42.81 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chr19_+_15094309 | 42.12 |
ENSRNOT00000083500
|
Ces1f
|
carboxylesterase 1F |
| chr9_+_4094995 | 41.77 |
ENSRNOT00000089450
|
LOC100910057
|
sulfotransferase 1C2-like |
| chrX_-_1848904 | 34.79 |
ENSRNOT00000010984
|
Rgn
|
regucalcin |
| chr14_+_22517774 | 32.71 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr8_+_115150514 | 32.06 |
ENSRNOT00000016504
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr5_+_133896141 | 32.04 |
ENSRNOT00000011434
|
Pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr17_+_76306585 | 31.53 |
ENSRNOT00000065978
|
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr18_-_15089988 | 29.73 |
ENSRNOT00000074116
|
Mep1b
|
meprin A subunit beta |
| chr5_-_159962218 | 27.37 |
ENSRNOT00000050729
|
Clcnkb
|
chloride voltage-gated channel Kb |
| chr3_-_46726946 | 27.07 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr7_-_68512397 | 26.91 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr2_+_68820615 | 26.86 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr8_+_85355766 | 25.25 |
ENSRNOT00000010583
|
Gcm1
|
glial cells missing homolog 1 |
| chr1_+_153589471 | 24.76 |
ENSRNOT00000023205
|
Fzd4
|
frizzled class receptor 4 |
| chr9_+_4107246 | 24.48 |
ENSRNOT00000078212
|
AABR07066160.1
|
|
| chr16_+_18690649 | 24.01 |
ENSRNOT00000015190
|
Mat1a
|
methionine adenosyltransferase 1A |
| chr10_-_47997097 | 24.00 |
ENSRNOT00000032092
|
Slc5a10
|
solute carrier family 5 member 10 |
| chr18_+_69841053 | 23.79 |
ENSRNOT00000071545
ENSRNOT00000030613 ENSRNOT00000075543 |
Mro
|
maestro |
| chr1_+_100164400 | 22.63 |
ENSRNOT00000080334
|
LOC108348116
|
glandular kallikrein-3, submandibular-like |
| chr13_+_55274199 | 21.76 |
ENSRNOT00000029679
|
Atp6v1g3
|
ATPase H+ transporting V1 subunit G3 |
| chr8_-_53816447 | 21.34 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr9_-_4327679 | 21.32 |
ENSRNOT00000073468
|
LOC100910235
|
sulfotransferase 1C1-like |
| chr9_+_95285592 | 20.65 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr3_-_2453415 | 20.63 |
ENSRNOT00000079773
|
Slc34a3
|
solute carrier family 34 member 3 |
| chr11_-_77703255 | 20.25 |
ENSRNOT00000083319
|
Cldn16
|
claudin 16 |
| chr8_+_80965255 | 20.16 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr19_+_39357791 | 20.02 |
ENSRNOT00000086435
ENSRNOT00000015006 |
Cyb5b
|
cytochrome b5 type B |
| chr12_-_41625403 | 19.88 |
ENSRNOT00000001876
|
Sds
|
serine dehydratase |
| chr2_-_117454769 | 19.22 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr20_-_27117663 | 19.11 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr14_-_44613904 | 18.80 |
ENSRNOT00000003811
|
Klb
|
klotho beta |
| chr3_-_51297852 | 18.39 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr10_-_59883839 | 17.75 |
ENSRNOT00000093579
|
Aspa
|
aspartoacylase |
| chr11_-_32508420 | 16.45 |
ENSRNOT00000002717
|
Kcne1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr8_+_49713190 | 16.11 |
ENSRNOT00000022074
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr2_+_128461224 | 16.04 |
ENSRNOT00000018872
|
Jade1
|
jade family PHD finger 1 |
| chr19_+_55982740 | 15.83 |
ENSRNOT00000021397
|
Dpep1
|
dipeptidase 1 (renal) |
| chr1_-_216663720 | 15.03 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr10_-_18506337 | 14.85 |
ENSRNOT00000043036
|
Gabrp
|
gamma-aminobutyric acid type A receptor pi subunit |
| chr19_+_60017746 | 14.48 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr5_+_137371825 | 14.01 |
ENSRNOT00000072816
|
Tmem125
|
transmembrane protein 125 |
| chr4_+_22084954 | 13.94 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr15_-_8989580 | 13.85 |
ENSRNOT00000061402
|
Thrb
|
thyroid hormone receptor beta |
| chr2_-_94315976 | 13.84 |
ENSRNOT00000071039
|
AABR07009357.1
|
|
| chr4_-_117607428 | 13.25 |
ENSRNOT00000021243
|
LOC103690139
|
probable N-acetyltransferase CML6 |
| chr1_+_14224393 | 13.05 |
ENSRNOT00000016037
|
Perp
|
PERP, TP53 apoptosis effector |
| chr7_+_101146820 | 12.58 |
ENSRNOT00000080206
|
AABR07058124.3
|
|
| chr1_+_15180328 | 12.51 |
ENSRNOT00000050656
|
Il20ra
|
interleukin 20 receptor subunit alpha |
| chr1_-_213907144 | 12.48 |
ENSRNOT00000054874
|
Sigirr
|
single Ig and TIR domain containing |
| chr13_-_37287458 | 11.99 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr5_-_57429245 | 11.63 |
ENSRNOT00000013803
|
Aqp3
|
aquaporin 3 |
| chr4_+_101180404 | 11.45 |
ENSRNOT00000007624
|
Suclg1
|
succinate-CoA ligase, alpha subunit |
| chr4_+_99063181 | 10.98 |
ENSRNOT00000008840
|
Fabp1
|
fatty acid binding protein 1 |
| chr4_-_23119005 | 10.89 |
ENSRNOT00000048061
|
Steap4
|
STEAP4 metalloreductase |
| chr1_+_141218095 | 10.82 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr7_-_75364166 | 10.65 |
ENSRNOT00000036852
|
Snx31
|
sorting nexin 31 |
| chr3_-_136936650 | 10.62 |
ENSRNOT00000036273
ENSRNOT00000083061 |
Kif16b
|
kinesin family member 16B |
| chr9_-_23352668 | 10.46 |
ENSRNOT00000075279
|
Mut
|
methylmalonyl CoA mutase |
| chr14_-_8600512 | 10.35 |
ENSRNOT00000092537
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_+_220432037 | 10.30 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr3_-_105214989 | 10.16 |
ENSRNOT00000037895
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr1_+_81372650 | 10.14 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chrX_+_34623405 | 10.11 |
ENSRNOT00000041924
|
Nhs
|
NHS actin remodeling regulator |
| chr11_-_32088002 | 9.92 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr1_+_190229528 | 9.79 |
ENSRNOT00000088121
|
Abca16
|
ATP-binding cassette, subfamily A (ABC1), member 16 |
| chr5_-_64718131 | 9.57 |
ENSRNOT00000088211
|
Acnat1
|
acyl-coenzyme A amino acid N-acyltransferase 1 |
| chr17_-_27602934 | 9.37 |
ENSRNOT00000079298
|
Rreb1
|
ras responsive element binding protein 1 |
| chr8_+_119261747 | 9.18 |
ENSRNOT00000079648
|
AC132539.1
|
|
| chr8_+_116332796 | 8.81 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr8_+_65733400 | 8.67 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr17_+_9797907 | 8.54 |
ENSRNOT00000021638
|
Lman2
|
lectin, mannose-binding 2 |
| chr7_-_117680004 | 8.28 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr2_+_93792601 | 8.23 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr7_+_130308532 | 8.18 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr16_+_8211420 | 7.93 |
ENSRNOT00000079609
|
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr13_-_97173469 | 7.85 |
ENSRNOT00000017784
|
Kif26b
|
kinesin family member 26B |
| chr10_+_53570989 | 7.75 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr16_-_47535358 | 7.69 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr3_-_113376751 | 7.59 |
ENSRNOT00000030019
|
Catsper2
|
cation channel, sperm associated 2 |
| chr19_-_50220455 | 7.56 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr6_+_98284170 | 7.46 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr18_+_63023930 | 7.46 |
ENSRNOT00000085655
|
Impa2
|
inositol monophosphatase 2 |
| chr1_+_101427195 | 7.41 |
ENSRNOT00000028271
|
Gys1
|
glycogen synthase 1 |
| chr1_-_199716205 | 7.30 |
ENSRNOT00000027290
|
RGD1310127
|
similar to cDNA sequence BC017158 |
| chr3_-_153952108 | 7.10 |
ENSRNOT00000050465
|
Adig
|
adipogenin |
| chr1_-_263959318 | 7.10 |
ENSRNOT00000068007
|
Pkd2l1
|
polycystin 2 like 1, transient receptor potential cation channel |
| chr12_+_13284532 | 7.08 |
ENSRNOT00000084045
|
Zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr3_+_60026747 | 7.07 |
ENSRNOT00000081881
|
Scrn3
|
secernin 3 |
| chr4_+_149261044 | 6.93 |
ENSRNOT00000066670
|
Cxcl12
|
C-X-C motif chemokine ligand 12 |
| chr5_-_168004724 | 6.86 |
ENSRNOT00000024711
|
Park7
|
Parkinsonism associated deglycase |
| chrX_+_54734385 | 6.77 |
ENSRNOT00000005023
|
Nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr2_-_211322719 | 6.74 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr2_-_124149621 | 6.66 |
ENSRNOT00000023448
ENSRNOT00000084825 ENSRNOT00000023437 |
Nudt6
|
nudix hydrolase 6 |
| chr3_-_156777999 | 6.56 |
ENSRNOT00000032588
ENSRNOT00000091208 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr5_-_173680290 | 6.45 |
ENSRNOT00000027564
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr9_+_23596964 | 6.39 |
ENSRNOT00000064279
|
LOC108351994
|
exocrine gland-secreted peptide 1-like |
| chr4_+_181315444 | 6.35 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr15_+_18540913 | 6.31 |
ENSRNOT00000010545
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr1_-_199655147 | 6.31 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chr6_+_99817431 | 6.26 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chr10_+_39435227 | 6.24 |
ENSRNOT00000042144
ENSRNOT00000077185 |
P4ha2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr15_-_4454958 | 6.19 |
ENSRNOT00000070933
|
Nudt13
|
nudix hydrolase 13 |
| chr10_-_4249739 | 6.17 |
ENSRNOT00000003150
|
Snx29
|
sorting nexin 29 |
| chr8_-_47339343 | 6.11 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr8_-_123371257 | 6.03 |
ENSRNOT00000017243
|
Stt3b
|
STT3B, catalytic subunit of the oligosaccharyltransferase complex |
| chr9_+_17728816 | 5.84 |
ENSRNOT00000065754
|
Capn11
|
calpain 11 |
| chr16_-_6077978 | 5.80 |
ENSRNOT00000020823
|
Il17rb
|
interleukin 17 receptor B |
| chr18_+_16146447 | 5.78 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr5_-_171327450 | 5.73 |
ENSRNOT00000088710
|
Ccdc27
|
coiled-coil domain containing 27 |
| chr1_+_267689328 | 5.73 |
ENSRNOT00000077738
|
Cfap58
|
cilia and flagella associated protein 58 |
| chr8_+_116054465 | 5.73 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr3_-_171893901 | 5.63 |
ENSRNOT00000091966
|
Apcdd1l
|
APC down-regulated 1 like |
| chr10_-_60275762 | 5.53 |
ENSRNOT00000074380
|
Olr1498
|
olfactory receptor 1498 |
| chr2_-_259382765 | 5.43 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr4_+_56337695 | 5.41 |
ENSRNOT00000071926
|
Lep
|
leptin |
| chr6_-_139973811 | 5.40 |
ENSRNOT00000082875
|
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr7_+_38858062 | 5.38 |
ENSRNOT00000006234
|
Kera
|
keratocan |
| chr1_+_202770775 | 5.37 |
ENSRNOT00000027704
|
Wdr11
|
WD repeat domain 11 |
| chr10_-_60126696 | 5.28 |
ENSRNOT00000026857
|
Olr1485
|
olfactory receptor 1485 |
| chr1_+_237574788 | 5.26 |
ENSRNOT00000042738
|
AABR07006504.1
|
|
| chr7_-_114590119 | 5.21 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr14_-_16903242 | 5.16 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr3_-_35801614 | 5.10 |
ENSRNOT00000066058
|
Mmadhc
|
methylmalonic aciduria and homocystinuria, cblD type |
| chr13_+_103268068 | 5.08 |
ENSRNOT00000088921
ENSRNOT00000003249 |
Bpnt1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr8_-_76452554 | 5.06 |
ENSRNOT00000079815
|
Gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr20_-_30947484 | 5.04 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr20_+_3553455 | 4.91 |
ENSRNOT00000090080
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chrX_-_147472677 | 4.67 |
ENSRNOT00000072063
|
Gm6760
|
predicted gene 6760 |
| chr4_-_165747201 | 4.66 |
ENSRNOT00000007435
|
Tas2r130
|
taste receptor, type 2, member 130 |
| chr14_-_85772762 | 4.54 |
ENSRNOT00000015627
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr11_-_65350442 | 4.53 |
ENSRNOT00000003773
|
Gpr156
|
G protein-coupled receptor 156 |
| chr7_-_73450262 | 4.48 |
ENSRNOT00000006943
|
Nipal2
|
NIPA-like domain containing 2 |
| chr6_-_71199110 | 4.30 |
ENSRNOT00000081883
|
Prkd1
|
protein kinase D1 |
| chr20_+_26755911 | 4.20 |
ENSRNOT00000083017
|
Herc4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr5_-_157573183 | 4.15 |
ENSRNOT00000064418
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr8_-_63750531 | 4.12 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr14_+_76866135 | 4.01 |
ENSRNOT00000037291
|
Zfp518b
|
zinc finger protein 518B |
| chr4_+_14212925 | 3.95 |
ENSRNOT00000076946
|
LOC103690020
|
platelet glycoprotein 4-like |
| chr5_+_79367663 | 3.92 |
ENSRNOT00000011508
|
Atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr1_-_72129269 | 3.85 |
ENSRNOT00000049179
|
Vom1r35
|
vomeronasal 1 receptor 35 |
| chr4_+_45000973 | 3.82 |
ENSRNOT00000084736
ENSRNOT00000080880 |
ST7
|
suppression of tumorigenicity 7 |
| chr13_+_100214353 | 3.77 |
ENSRNOT00000073693
|
LOC686031
|
hypothetical protein LOC686031 |
| chr19_-_30821999 | 3.75 |
ENSRNOT00000072648
|
Gm8797
|
predicted pseudogene 8797 |
| chr3_-_76187045 | 3.72 |
ENSRNOT00000075650
|
LOC686683
|
similar to olfactory receptor 73 |
| chr10_-_46582854 | 3.72 |
ENSRNOT00000004753
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr20_-_831447 | 3.69 |
ENSRNOT00000000976
|
Olr1693
|
olfactory receptor 1693 |
| chr10_+_109893700 | 3.67 |
ENSRNOT00000082551
|
Lrrc45
|
leucine rich repeat containing 45 |
| chr19_+_38768467 | 3.60 |
ENSRNOT00000027346
|
Cdh1
|
cadherin 1 |
| chr10_+_92769031 | 3.59 |
ENSRNOT00000090759
|
AABR07030543.1
|
|
| chr12_+_44046454 | 3.57 |
ENSRNOT00000064756
|
Fbxw8
|
F-box and WD repeat domain containing 8 |
| chr14_-_6148830 | 3.57 |
ENSRNOT00000075091
ENSRNOT00000092683 |
LOC100366030
|
rCG37858-like |
| chr14_+_22251499 | 3.54 |
ENSRNOT00000087991
ENSRNOT00000002705 |
Ugt2a1
|
UDP glucuronosyltransferase 2 family, polypeptide A1 |
| chr1_+_267618565 | 3.49 |
ENSRNOT00000076251
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr1_-_190965115 | 3.48 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr10_-_46206135 | 3.47 |
ENSRNOT00000091471
|
Cops3
|
COP9 signalosome subunit 3 |
| chr1_+_172822544 | 3.42 |
ENSRNOT00000090018
|
AABR07004960.1
|
|
| chr3_-_21027947 | 3.41 |
ENSRNOT00000051973
|
Olr421
|
olfactory receptor 421 |
| chr5_-_164714145 | 3.39 |
ENSRNOT00000055680
|
LOC100911485
|
mitofusin-2-like |
| chr19_-_25961666 | 3.39 |
ENSRNOT00000004091
|
Calr
|
calreticulin |
| chr13_-_88307988 | 3.38 |
ENSRNOT00000003812
|
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr8_-_43524220 | 3.35 |
ENSRNOT00000088755
|
Olr1315
|
olfactory receptor 1315 |
| chr13_+_52889737 | 3.35 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr10_+_49368314 | 3.33 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr7_-_49732764 | 3.30 |
ENSRNOT00000006453
|
Myf5
|
myogenic factor 5 |
| chr8_-_116764780 | 3.25 |
ENSRNOT00000026216
|
Fam212a
|
family with sequence similarity 212, member A |
| chr20_-_2913785 | 3.23 |
ENSRNOT00000088584
|
Btnl7
|
butyrophilin-like 7 |
| chrX_+_72685300 | 3.22 |
ENSRNOT00000076215
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr6_-_122239614 | 3.21 |
ENSRNOT00000005015
|
Galc
|
galactosylceramidase |
| chr9_-_63291350 | 3.21 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr7_-_117289961 | 3.12 |
ENSRNOT00000042642
|
Plec
|
plectin |
| chr1_+_264260505 | 3.09 |
ENSRNOT00000018815
|
Wnt8b
|
wingless-type MMTV integration site family, member 8B |
| chr1_-_229804614 | 3.02 |
ENSRNOT00000049665
|
Olr343
|
olfactory receptor 343 |
| chr5_-_75057752 | 2.96 |
ENSRNOT00000016447
|
Txn1
|
thioredoxin 1 |
| chr5_+_172648950 | 2.96 |
ENSRNOT00000055361
|
Faap20
|
Fanconi anemia core complex associated protein 20 |
| chr8_-_42979305 | 2.88 |
ENSRNOT00000073449
|
LOC100910439
|
olfactory receptor 146-like |
| chr17_-_15519060 | 2.85 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr3_-_73493621 | 2.75 |
ENSRNOT00000071782
|
Olr482
|
olfactory receptor 482 |
| chr6_-_104290579 | 2.75 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr3_+_77072410 | 2.72 |
ENSRNOT00000078540
|
Olr649
|
olfactory receptor 649 |
| chr20_+_734642 | 2.72 |
ENSRNOT00000073543
|
Olr1688
|
olfactory receptor 1688 |
| chr3_-_165537940 | 2.71 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr2_-_29121104 | 2.70 |
ENSRNOT00000020543
|
Tnpo1
|
transportin 1 |
| chrX_+_106230013 | 2.65 |
ENSRNOT00000067828
|
AABR07040617.1
|
|
| chr2_-_123851854 | 2.64 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr10_+_14481393 | 2.64 |
ENSRNOT00000091072
|
Ptx4
|
pentraxin 4 |
| chr10_+_55997614 | 2.60 |
ENSRNOT00000013071
|
Naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr15_+_108453147 | 2.59 |
ENSRNOT00000018486
|
Timm8a2
|
translocase of inner mitochondrial membrane 8A2 |
| chrX_+_115721251 | 2.57 |
ENSRNOT00000060090
|
Trpc5os
|
TRPC5 opposite strand |
| chr3_-_76279104 | 2.56 |
ENSRNOT00000007796
|
Olr608
|
olfactory receptor 608 |
| chr5_-_58288125 | 2.53 |
ENSRNOT00000068752
|
Fam205a
|
family with sequence similarity 205, member A |
| chr1_-_149778698 | 2.50 |
ENSRNOT00000089920
|
LOC100909935
|
olfactory receptor 6F1-like |
| chr4_+_71512695 | 2.50 |
ENSRNOT00000037220
|
Tas2r139
|
taste receptor, type 2, member 139 |
| chr1_+_207508414 | 2.48 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr2_-_35435719 | 2.48 |
ENSRNOT00000087827
|
LOC100910041
|
olfactory receptor 146-like |
| chr6_-_142385773 | 2.47 |
ENSRNOT00000071555
|
AABR07065814.7
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Meis1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.6 | 34.8 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 6.4 | 19.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 5.4 | 27.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 5.4 | 26.9 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 5.1 | 25.3 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 5.0 | 19.9 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 4.8 | 24.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 4.3 | 68.4 | GO:0051923 | sulfation(GO:0051923) |
| 4.2 | 12.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 4.1 | 24.8 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 4.0 | 32.3 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 3.7 | 11.0 | GO:0051977 | lysophospholipid transport(GO:0051977) |
| 3.4 | 27.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 2.9 | 20.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 2.9 | 11.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 2.8 | 13.9 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 2.8 | 8.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 2.6 | 84.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 2.5 | 10.2 | GO:1900158 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 2.5 | 15.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 2.5 | 9.9 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 2.4 | 14.5 | GO:0003383 | apical constriction(GO:0003383) |
| 2.3 | 18.8 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 2.3 | 9.4 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 2.3 | 11.6 | GO:0015793 | glycerol transport(GO:0015793) |
| 2.3 | 83.7 | GO:0017144 | drug metabolic process(GO:0017144) |
| 2.3 | 13.9 | GO:0008050 | female courtship behavior(GO:0008050) |
| 2.3 | 20.6 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 2.3 | 6.9 | GO:1903122 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 2.3 | 20.3 | GO:0070633 | transepithelial transport(GO:0070633) |
| 2.2 | 8.8 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 2.1 | 19.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 2.0 | 7.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 1.8 | 23.5 | GO:0071468 | cellular response to acidic pH(GO:0071468) |
| 1.7 | 6.9 | GO:1990478 | response to ultrasound(GO:1990478) |
| 1.7 | 5.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 1.6 | 1.6 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 1.5 | 16.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 1.4 | 37.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 1.4 | 10.9 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 1.3 | 5.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 1.2 | 18.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.2 | 4.9 | GO:0060066 | oviduct development(GO:0060066) |
| 1.2 | 13.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 1.2 | 5.8 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 1.2 | 5.8 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 1.1 | 44.7 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 1.1 | 3.4 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 1.1 | 20.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 1.0 | 6.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 1.0 | 10.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 1.0 | 17.2 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.9 | 7.5 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.9 | 17.7 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.9 | 3.5 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.8 | 8.9 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.8 | 2.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.8 | 2.3 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.7 | 8.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.7 | 4.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.7 | 7.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.7 | 5.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.7 | 2.6 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.6 | 4.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.6 | 8.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.6 | 1.7 | GO:0048014 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.6 | 5.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.6 | 6.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.5 | 6.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 | 3.2 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.5 | 9.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.5 | 1.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.5 | 3.5 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.5 | 2.5 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.5 | 6.8 | GO:0030238 | male sex determination(GO:0030238) |
| 0.4 | 3.7 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.4 | 3.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 6.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.4 | 1.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.4 | 2.7 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.4 | 3.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 1.5 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.4 | 1.9 | GO:0032960 | regulation of inositol trisphosphate biosynthetic process(GO:0032960) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) somatotropin secreting cell differentiation(GO:0060126) |
| 0.4 | 8.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.4 | 13.2 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.4 | 7.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.3 | 4.5 | GO:0015693 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.3 | 3.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 7.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 3.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 8.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.3 | 4.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.3 | 1.4 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.3 | 0.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 5.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 8.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 8.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 10.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 3.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 4.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 2.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 1.0 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 | 3.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 10.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.2 | 18.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.2 | 7.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 2.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 5.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 1.9 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 11.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 8.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 1.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 1.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.9 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 1.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 2.8 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 2.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 5.2 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 1.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.1 | 2.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 83.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 0.2 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) |
| 0.1 | 0.3 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 1.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 7.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 3.1 | GO:0015918 | sterol transport(GO:0015918) |
| 0.0 | 6.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.7 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 17.0 | GO:0055114 | oxidation-reduction process(GO:0055114) |
| 0.0 | 1.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 1.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 31.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 4.0 | 12.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.9 | 8.8 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 2.4 | 14.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.7 | 8.7 | GO:0043293 | apoptosome(GO:0043293) |
| 1.6 | 6.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 1.4 | 24.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 1.3 | 16.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.3 | 11.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 1.1 | 11.0 | GO:0045179 | apical cortex(GO:0045179) |
| 1.1 | 7.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.8 | 3.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.8 | 22.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.7 | 6.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.7 | 6.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.6 | 2.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.6 | 17.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.6 | 3.6 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) 3M complex(GO:1990393) |
| 0.6 | 20.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.6 | 42.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.5 | 1.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.5 | 29.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 2.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 3.6 | GO:0043219 | lateral loop(GO:0043219) |
| 0.3 | 18.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 20.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 20.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 8.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 1.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 3.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 3.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 8.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 24.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.2 | 4.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 13.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 3.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 19.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 16.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 23.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 6.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 8.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 4.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 7.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 2.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 15.6 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 77.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 5.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 13.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 2.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 9.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 5.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 17.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 163.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 5.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 54.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 4.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 20.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 79.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 3.2 | GO:0030054 | cell junction(GO:0030054) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.5 | 31.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 8.0 | 24.0 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 6.6 | 19.9 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 5.9 | 17.7 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 5.6 | 67.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 5.4 | 26.9 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 5.3 | 15.8 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 4.6 | 13.9 | GO:0008458 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 4.2 | 12.5 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 4.1 | 20.6 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 3.9 | 11.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 3.8 | 11.5 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 3.0 | 21.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 3.0 | 51.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 2.7 | 24.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 2.6 | 84.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 2.6 | 10.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 2.3 | 16.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 2.3 | 6.9 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 2.2 | 10.9 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.0 | 10.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 1.9 | 7.5 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 1.8 | 56.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.8 | 11.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.8 | 8.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.7 | 6.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 1.6 | 6.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.6 | 6.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 1.4 | 27.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 1.3 | 10.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.3 | 6.4 | GO:0042731 | PH domain binding(GO:0042731) |
| 1.3 | 5.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) inositol bisphosphate phosphatase activity(GO:0016312) |
| 1.3 | 5.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.3 | 13.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.2 | 3.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 1.2 | 17.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.1 | 4.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.1 | 3.4 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.1 | 5.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 15.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.1 | 7.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.9 | 3.5 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.9 | 6.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.8 | 46.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.7 | 5.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.7 | 26.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.6 | 7.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.6 | 8.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.6 | 18.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.5 | 2.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 6.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.5 | 2.6 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.5 | 1.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.5 | 5.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.5 | 8.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.5 | 11.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.5 | 9.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 14.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.5 | 2.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.5 | 34.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.5 | 73.8 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.4 | 8.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 5.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 1.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 8.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 1.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.4 | 4.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 4.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 17.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.4 | 3.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 1.3 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.3 | 1.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 20.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.3 | 4.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 7.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.3 | 30.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 5.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 7.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 27.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 1.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 4.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 3.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 5.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 4.9 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.2 | 1.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.2 | 2.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 4.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 0.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 9.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 56.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 13.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 2.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 3.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.3 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 11.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.0 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.1 | 7.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 6.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.0 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 3.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 8.0 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.0 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 4.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 3.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 26.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 5.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 1.4 | 26.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 1.3 | 27.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.9 | 24.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.5 | 5.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.5 | 35.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 6.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 12.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 13.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 18.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 7.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 13.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 6.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 4.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 10.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 6.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 8.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 5.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 26.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 1.3 | 26.9 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 1.3 | 67.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 1.3 | 24.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.0 | 33.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 1.0 | 11.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 24.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.8 | 24.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.7 | 10.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.7 | 18.8 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.6 | 11.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.6 | 13.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.6 | 9.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.5 | 8.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 6.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 4.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.4 | 5.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 5.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.4 | 5.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.4 | 5.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 4.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 10.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 16.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 4.7 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.2 | 5.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 6.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 19.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 6.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 6.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 7.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 15.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 7.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 11.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 5.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 4.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |