Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Mef2d_Mef2a
Z-value: 6.11
Transcription factors associated with Mef2d_Mef2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
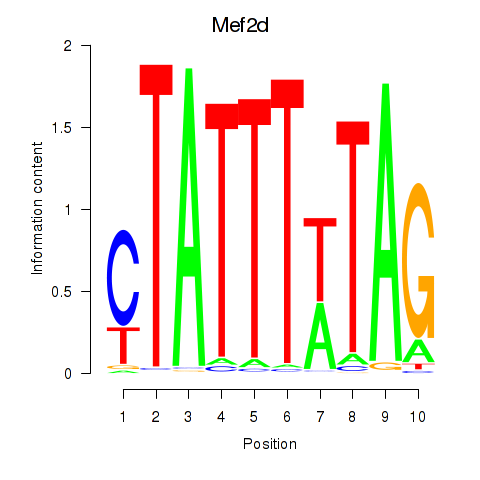
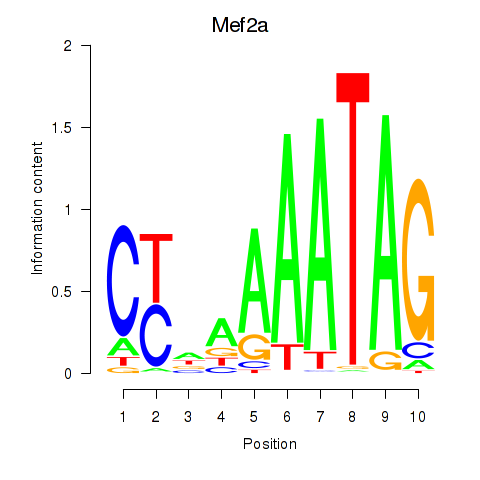
Mef2d
|
ENSRNOG00000031778 | myocyte enhancer factor 2D |
|
Mef2a
|
ENSRNOG00000047756 | myocyte enhancer factor 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2a | rn6_v1_chr1_-_128287151_128287151 | 0.55 | 3.7e-27 | Click! |
| Mef2d | rn6_v1_chr2_+_187512164_187512164 | 0.41 | 2.1e-14 | Click! |
Activity profile of Mef2d_Mef2a motif
Sorted Z-values of Mef2d_Mef2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_41212072 | 744.73 |
ENSRNOT00000085596
|
Ppp1r3a
|
protein phosphatase 1, regulatory subunit 3A |
| chr1_-_199624783 | 743.53 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr3_+_53563194 | 694.18 |
ENSRNOT00000048300
|
Xirp2
|
xin actin-binding repeat containing 2 |
| chr1_-_25839198 | 642.18 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr1_-_277181345 | 562.02 |
ENSRNOT00000038017
ENSRNOT00000038038 |
Nrap
|
nebulin-related anchoring protein |
| chr10_+_86337728 | 562.02 |
ENSRNOT00000085408
|
Tcap
|
titin-cap |
| chr9_-_73948583 | 547.50 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr10_+_54352270 | 524.23 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr2_-_45518502 | 472.78 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr9_+_47536824 | 462.07 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr1_-_104166367 | 452.35 |
ENSRNOT00000092211
|
Csrp3
|
cysteine and glycine rich protein 3 |
| chr2_+_205568935 | 445.78 |
ENSRNOT00000025248
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr11_+_66713888 | 445.69 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr15_+_4064706 | 416.93 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chrX_-_40086870 | 416.06 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr1_+_215609036 | 398.66 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr1_-_143398093 | 374.57 |
ENSRNOT00000078916
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr16_-_10941414 | 371.74 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr8_+_110982777 | 370.21 |
ENSRNOT00000010992
|
Ky
|
kyphoscoliosis peptidase |
| chr10_-_56558487 | 369.95 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr7_-_118108864 | 369.94 |
ENSRNOT00000006184
|
Mb
|
myoglobin |
| chr3_+_55910177 | 360.68 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr2_-_227207584 | 347.06 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr2_+_204512302 | 344.54 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr10_+_70262361 | 343.45 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr2_+_219598162 | 335.29 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr10_+_53740841 | 329.48 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr5_-_22769907 | 323.28 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chrX_-_64726210 | 319.01 |
ENSRNOT00000076012
ENSRNOT00000086265 |
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr1_+_142883040 | 315.29 |
ENSRNOT00000015898
|
Alpk3
|
alpha-kinase 3 |
| chr10_-_32471454 | 312.12 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr7_-_98197087 | 310.23 |
ENSRNOT00000010484
ENSRNOT00000079961 |
Klhl38
|
kelch-like family member 38 |
| chr1_+_199449973 | 309.38 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
| chr6_+_73553210 | 302.80 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr10_+_53818818 | 287.71 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr4_+_71675383 | 285.29 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr10_+_53778662 | 282.31 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr3_+_112228919 | 273.35 |
ENSRNOT00000011761
|
Capn3
|
calpain 3 |
| chr1_-_198233588 | 270.40 |
ENSRNOT00000088473
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr1_+_215609645 | 262.96 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr12_+_49761120 | 256.19 |
ENSRNOT00000070961
|
Myo18b
|
myosin XVIIIb |
| chr13_-_90602365 | 256.07 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr5_+_90338795 | 251.60 |
ENSRNOT00000077864
ENSRNOT00000058882 |
LOC298139
|
similar to RIKEN cDNA 2310003M01 |
| chr1_-_198233215 | 245.39 |
ENSRNOT00000087928
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr8_+_119030875 | 240.31 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr1_+_31531143 | 235.91 |
ENSRNOT00000034704
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr3_+_112228720 | 232.01 |
ENSRNOT00000079079
|
Capn3
|
calpain 3 |
| chr4_+_7158448 | 230.15 |
ENSRNOT00000076953
|
Asb10
|
ankyrin repeat and SOCS box-containing 10 |
| chr3_+_79940561 | 225.19 |
ENSRNOT00000016652
|
Mybpc3
|
myosin binding protein C, cardiac |
| chr1_+_221756286 | 221.27 |
ENSRNOT00000028636
|
Pygm
|
glycogen phosphorylase, muscle associated |
| chr10_+_11240138 | 214.06 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr5_+_152533349 | 213.82 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr13_+_52624878 | 207.21 |
ENSRNOT00000076054
ENSRNOT00000076299 |
Tnni1
|
troponin I1, slow skeletal type |
| chr16_+_49266903 | 206.55 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chrX_-_31780425 | 205.14 |
ENSRNOT00000004693
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr1_+_167197549 | 200.85 |
ENSRNOT00000027427
|
Art1
|
ADP-ribosyltransferase 1 |
| chr15_-_27819376 | 198.41 |
ENSRNOT00000067400
|
A930018M24Rik
|
RIKEN cDNA A930018M24 gene |
| chr10_-_8654892 | 196.46 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr16_+_2537248 | 194.62 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr9_+_82571269 | 190.45 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr5_+_173640780 | 190.20 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr9_-_97151832 | 189.12 |
ENSRNOT00000040169
|
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr2_+_119197239 | 189.05 |
ENSRNOT00000048030
|
Usp13
|
ubiquitin specific peptidase 13 |
| chr17_-_78735324 | 178.11 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chrX_+_71342775 | 175.98 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr8_+_55178289 | 174.46 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr15_+_2766710 | 172.40 |
ENSRNOT00000017483
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr8_-_33017854 | 172.11 |
ENSRNOT00000011386
|
Barx2
|
BARX homeobox 2 |
| chr10_-_82785142 | 171.95 |
ENSRNOT00000005381
|
Sgca
|
sarcoglycan, alpha |
| chr2_-_172361779 | 171.94 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr17_+_32973695 | 170.74 |
ENSRNOT00000065674
|
RGD1565323
|
similar to OTTMUSP00000000621 |
| chr2_+_198655437 | 168.15 |
ENSRNOT00000028781
|
Hfe2
|
hemochromatosis type 2 (juvenile) |
| chr16_-_73827488 | 167.18 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chrX_-_157013443 | 161.04 |
ENSRNOT00000082711
|
Srpk3
|
SRSF protein kinase 3 |
| chr18_+_32273770 | 160.41 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr2_+_104416972 | 158.97 |
ENSRNOT00000017125
|
Trim55
|
tripartite motif-containing 55 |
| chr8_+_130416355 | 157.32 |
ENSRNOT00000026234
|
Klhl40
|
kelch-like family member 40 |
| chr9_+_82556573 | 157.29 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr13_-_90676629 | 154.79 |
ENSRNOT00000058143
|
Atp1a2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr3_+_58965552 | 152.36 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr8_-_55177818 | 151.99 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr6_+_56625650 | 149.19 |
ENSRNOT00000008803
|
Meox2
|
mesenchyme homeobox 2 |
| chr9_+_98490608 | 146.17 |
ENSRNOT00000027232
|
Klhl30
|
kelch-like family member 30 |
| chr2_-_104958034 | 144.69 |
ENSRNOT00000080699
|
Gyg1
|
glycogenin 1 |
| chr14_-_17171580 | 144.58 |
ENSRNOT00000085525
|
Art3
|
ADP-ribosyltransferase 3 |
| chr7_+_70807867 | 135.97 |
ENSRNOT00000010639
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr4_-_99125111 | 132.41 |
ENSRNOT00000009184
|
Smyd1
|
SET and MYND domain containing 1 |
| chr2_-_247988462 | 131.47 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chr2_-_227411964 | 125.51 |
ENSRNOT00000019931
|
Synpo2
|
synaptopodin 2 |
| chr12_-_23841049 | 124.83 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr1_+_101397828 | 124.17 |
ENSRNOT00000028189
|
Kcna7
|
potassium voltage-gated channel subfamily A member 7 |
| chr15_-_28314459 | 119.77 |
ENSRNOT00000042055
ENSRNOT00000040540 |
Ndrg2
|
NDRG family member 2 |
| chr2_+_196334626 | 112.03 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr7_-_49741540 | 110.27 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr1_-_264975132 | 110.01 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr15_+_86243148 | 109.59 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr4_-_61720956 | 109.12 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr15_-_37325178 | 107.23 |
ENSRNOT00000011699
|
Gja3
|
gap junction protein, alpha 3 |
| chr13_+_51034256 | 106.65 |
ENSRNOT00000004528
ENSRNOT00000046854 ENSRNOT00000087320 |
Mybph
|
myosin binding protein H |
| chr1_-_89488223 | 101.81 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr10_+_39655455 | 96.30 |
ENSRNOT00000058817
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr14_-_43072843 | 92.82 |
ENSRNOT00000064263
|
Limch1
|
LIM and calponin homology domains 1 |
| chr13_+_52662996 | 91.18 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr4_+_85551502 | 90.49 |
ENSRNOT00000087191
ENSRNOT00000015692 |
Aqp1
|
aquaporin 1 |
| chr8_-_109560747 | 89.89 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr10_-_89338739 | 80.98 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr5_+_154598758 | 79.54 |
ENSRNOT00000015776
|
Tcea3
|
transcription elongation factor A3 |
| chr2_-_198016898 | 74.55 |
ENSRNOT00000025523
|
Car14
|
carbonic anhydrase 14 |
| chr9_-_19372673 | 71.11 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr9_-_50762082 | 68.39 |
ENSRNOT00000015492
|
Mettl21c
|
methyltransferase like 21C |
| chr6_-_137733026 | 67.97 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr4_+_144192989 | 65.17 |
ENSRNOT00000007523
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr17_+_72218769 | 64.88 |
ENSRNOT00000041346
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr19_+_52313795 | 64.87 |
ENSRNOT00000021630
|
Wfdc1
|
WAP four-disulfide core domain 1 |
| chr2_-_261337163 | 63.73 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chr2_+_93792601 | 63.70 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr9_-_9702306 | 63.18 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr6_-_136550371 | 60.24 |
ENSRNOT00000065971
|
Rd3l
|
retinal degeneration 3-like |
| chr5_-_144274981 | 57.03 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr13_-_81214494 | 56.62 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr20_+_3830164 | 56.32 |
ENSRNOT00000045533
ENSRNOT00000084117 |
Col11a2
|
collagen type XI alpha 2 chain |
| chr14_+_63095720 | 55.16 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr1_-_24056373 | 52.91 |
ENSRNOT00000015566
|
Slc2a12
|
solute carrier family 2 member 12 |
| chr1_-_56683731 | 45.14 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr4_+_148782479 | 42.96 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr11_+_42945084 | 41.88 |
ENSRNOT00000002292
|
Crybg3
|
crystallin beta-gamma domain containing 3 |
| chr12_+_17734133 | 41.16 |
ENSRNOT00000042117
|
Pdgfa
|
platelet derived growth factor subunit A |
| chr2_+_189997129 | 39.97 |
ENSRNOT00000015958
|
S100a4
|
S100 calcium-binding protein A4 |
| chr3_-_64024205 | 38.69 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr1_-_142164263 | 38.32 |
ENSRNOT00000016281
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr8_-_127782070 | 38.32 |
ENSRNOT00000045493
|
Plcd1
|
phospholipase C, delta 1 |
| chrM_+_7006 | 37.61 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr1_-_142164007 | 36.30 |
ENSRNOT00000078982
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr2_-_197991198 | 34.18 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr3_+_63353178 | 34.17 |
ENSRNOT00000091211
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr3_-_161299024 | 33.66 |
ENSRNOT00000021216
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr3_+_151126591 | 32.26 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr7_+_142905758 | 28.17 |
ENSRNOT00000078663
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr4_+_159563798 | 26.74 |
ENSRNOT00000081426
|
Fgf6
|
fibroblast growth factor 6 |
| chr9_-_52238564 | 26.40 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr4_+_89079014 | 26.18 |
ENSRNOT00000087451
|
Herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr2_-_197991574 | 25.17 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr1_+_238222521 | 23.70 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr10_-_86688730 | 22.79 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr13_-_1946508 | 18.77 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr11_-_60679555 | 15.43 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr8_-_120446455 | 14.50 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr20_+_20236151 | 13.64 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr9_+_47134034 | 13.31 |
ENSRNOT00000020108
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr7_+_132857628 | 13.19 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr2_+_84645084 | 12.71 |
ENSRNOT00000015448
|
Cmbl
|
carboxymethylenebutenolidase homolog |
| chr6_+_8284878 | 11.41 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr4_-_85329362 | 11.11 |
ENSRNOT00000014925
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr3_+_65816569 | 10.81 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr5_-_107039925 | 8.39 |
ENSRNOT00000049705
|
Ifna5
|
interferon, alpha 5 |
| chr11_-_28777603 | 7.11 |
ENSRNOT00000002132
|
Krtap14
|
keratin associated protein 14 |
| chr3_+_60026747 | 6.76 |
ENSRNOT00000081881
|
Scrn3
|
secernin 3 |
| chr2_+_189993262 | 6.52 |
ENSRNOT00000016034
|
S100a3
|
S100 calcium binding protein A3 |
| chr10_+_55687050 | 5.20 |
ENSRNOT00000057136
|
Per1
|
period circadian clock 1 |
| chr7_-_60743328 | 4.72 |
ENSRNOT00000066767
|
Mdm2
|
MDM2 proto-oncogene |
| chr6_-_33691301 | 4.53 |
ENSRNOT00000008008
|
Rhob
|
ras homolog family member B |
| chrX_-_112473822 | 3.92 |
ENSRNOT00000079180
|
Col4a6
|
collagen type IV alpha 6 chain |
| chr2_-_187854363 | 3.89 |
ENSRNOT00000092993
|
Lmna
|
lamin A/C |
| chr2_+_203200427 | 3.26 |
ENSRNOT00000020566
|
Vtcn1
|
V-set domain containing T cell activation inhibitor 1 |
| chr1_+_78800754 | 1.31 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr13_-_70174565 | 1.16 |
ENSRNOT00000067135
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr2_+_184230459 | 1.06 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr7_-_132143470 | 1.03 |
ENSRNOT00000038946
ENSRNOT00000044092 ENSRNOT00000066528 ENSRNOT00000045553 ENSRNOT00000046744 ENSRNOT00000055742 |
Kif21a
|
kinesin family member 21A |
| chr3_-_2689084 | 0.54 |
ENSRNOT00000020926
|
Ptgds
|
prostaglandin D2 synthase |
| chr1_+_190671696 | 0.51 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr16_-_20994379 | 0.30 |
ENSRNOT00000027639
|
Tmem161a
|
transmembrane protein 161A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2d_Mef2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 253.6 | 1014.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 214.1 | 642.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 168.5 | 505.4 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 123.3 | 369.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 76.8 | 921.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 75.7 | 302.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 74.3 | 445.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 71.4 | 214.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 71.3 | 213.8 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 64.7 | 323.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 64.0 | 256.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 62.0 | 743.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 60.5 | 241.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 57.1 | 399.7 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 51.6 | 206.5 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 47.3 | 189.0 | GO:0044313 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) protein K6-linked deubiquitination(GO:0044313) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 41.6 | 124.8 | GO:0071348 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 38.7 | 154.8 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 37.9 | 378.9 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 36.9 | 331.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 36.8 | 515.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 36.4 | 109.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 33.9 | 101.8 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 32.8 | 557.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 32.1 | 96.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 32.1 | 160.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 31.5 | 157.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 30.2 | 90.5 | GO:0072237 | metanephric proximal tubule development(GO:0072237) |
| 29.9 | 119.8 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 29.8 | 149.2 | GO:0001757 | somite specification(GO:0001757) |
| 27.6 | 524.2 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 25.8 | 1004.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 25.6 | 256.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 23.8 | 285.3 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 21.8 | 174.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 18.5 | 166.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 18.4 | 110.3 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 17.4 | 715.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 17.0 | 68.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 15.6 | 171.9 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 15.6 | 934.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 14.2 | 370.0 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 14.1 | 704.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 13.7 | 41.2 | GO:1990401 | regulation of branching involved in salivary gland morphogenesis by epithelial-mesenchymal signaling(GO:0060683) embryonic lung development(GO:1990401) |
| 13.3 | 345.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 12.8 | 38.3 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 11.6 | 221.3 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 11.3 | 56.3 | GO:0060023 | soft palate development(GO:0060023) |
| 11.2 | 168.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 9.5 | 190.2 | GO:0014850 | response to muscle activity(GO:0014850) |
| 9.5 | 198.8 | GO:0001502 | cartilage condensation(GO:0001502) |
| 9.4 | 958.8 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 9.3 | 65.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 8.6 | 171.9 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 8.3 | 396.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 8.1 | 515.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 7.6 | 22.8 | GO:0060086 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 7.4 | 132.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 7.1 | 312.1 | GO:0055001 | muscle cell development(GO:0055001) |
| 6.8 | 74.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 6.6 | 26.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 6.3 | 157.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 5.5 | 71.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 5.4 | 178.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 5.4 | 167.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 5.2 | 343.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 4.6 | 63.7 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 4.4 | 13.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 4.2 | 152.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 3.8 | 18.8 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 3.4 | 23.7 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 3.4 | 161.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 2.8 | 11.1 | GO:2000292 | gastric motility(GO:0035482) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 2.7 | 59.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 2.7 | 239.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 2.6 | 1935.4 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 2.3 | 107.2 | GO:0009268 | response to pH(GO:0009268) |
| 2.3 | 64.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 1.7 | 5.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 1.5 | 49.0 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 1.5 | 172.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 1.3 | 38.7 | GO:0051642 | centrosome localization(GO:0051642) |
| 1.3 | 79.5 | GO:0006414 | translational elongation(GO:0006414) |
| 1.2 | 64.9 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 1.2 | 36.3 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 1.1 | 3.3 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.8 | 37.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.8 | 8.4 | GO:0033139 | regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033139) positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) serine phosphorylation of STAT protein(GO:0042501) |
| 0.5 | 4.5 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.3 | 57.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.3 | 3.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 15.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.5 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 57.0 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.1 | 1.3 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 315.0 | 945.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 202.3 | 1011.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 174.7 | 524.2 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 150.2 | 600.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 96.5 | 868.8 | GO:0005861 | troponin complex(GO:0005861) |
| 89.5 | 805.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 80.7 | 484.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 68.0 | 611.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 48.8 | 781.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 47.3 | 1324.8 | GO:0031430 | M band(GO:0031430) |
| 36.0 | 287.7 | GO:0032982 | myosin filament(GO:0032982) |
| 32.1 | 4820.2 | GO:0031674 | I band(GO:0031674) |
| 29.4 | 323.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 28.5 | 370.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 28.0 | 168.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 22.6 | 90.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 21.8 | 109.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 18.8 | 56.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 13.0 | 508.6 | GO:0016459 | myosin complex(GO:0016459) |
| 12.9 | 154.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 8.1 | 64.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 8.0 | 458.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 6.6 | 26.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 6.1 | 456.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 5.8 | 221.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 5.1 | 107.2 | GO:0005922 | connexon complex(GO:0005922) |
| 3.5 | 89.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 3.3 | 13.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 2.4 | 336.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 2.4 | 63.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 2.0 | 34.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 2.0 | 45.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 1.9 | 204.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 1.9 | 302.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 1.3 | 331.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 1.3 | 124.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 1.1 | 109.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 1.0 | 11.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.0 | 63.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.9 | 177.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.9 | 336.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.9 | 15.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.8 | 81.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.5 | 37.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.5 | 3.9 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.5 | 3.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 39.4 | GO:0016605 | PML body(GO:0016605) |
| 0.4 | 117.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.4 | 65.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 4.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 26.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 22.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 69.3 | GO:0005856 | cytoskeleton(GO:0005856) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 181.8 | 909.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 113.1 | 452.3 | GO:0061629 | telethonin binding(GO:0031433) RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 111.4 | 445.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 94.5 | 661.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 86.9 | 1391.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 73.8 | 221.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 73.7 | 515.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 65.4 | 719.2 | GO:0031432 | titin binding(GO:0031432) |
| 47.3 | 189.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 45.6 | 91.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 43.2 | 345.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 41.1 | 370.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 33.6 | 302.8 | GO:0043495 | protein anchor(GO:0043495) |
| 33.4 | 167.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 31.9 | 63.7 | GO:0031013 | troponin I binding(GO:0031013) |
| 28.0 | 168.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 26.0 | 781.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 24.7 | 369.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 23.9 | 694.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 22.9 | 206.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 20.7 | 144.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 20.4 | 448.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 18.1 | 90.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 16.1 | 241.1 | GO:0042805 | actinin binding(GO:0042805) |
| 15.5 | 154.8 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 15.0 | 285.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 14.8 | 326.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 13.6 | 109.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 12.4 | 248.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 8.9 | 160.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 8.8 | 343.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 8.6 | 112.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 8.3 | 240.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 8.0 | 96.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 7.9 | 23.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 7.7 | 107.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 6.6 | 262.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 6.3 | 18.8 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 6.2 | 390.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 5.9 | 152.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 5.7 | 74.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 5.0 | 40.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 4.5 | 251.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 4.4 | 13.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 4.4 | 13.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 3.9 | 1852.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 3.8 | 101.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 3.7 | 74.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 3.6 | 79.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 3.6 | 1282.8 | GO:0003779 | actin binding(GO:0003779) |
| 3.5 | 38.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 3.3 | 172.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 3.3 | 515.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 3.2 | 64.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 2.8 | 11.1 | GO:0043404 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 2.7 | 37.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 2.6 | 55.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 2.6 | 36.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 2.4 | 257.2 | GO:0003774 | motor activity(GO:0003774) |
| 2.0 | 1375.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 2.0 | 86.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 1.7 | 66.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 1.5 | 64.6 | GO:0070888 | E-box binding(GO:0070888) |
| 1.5 | 22.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 1.4 | 64.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 1.1 | 179.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 1.1 | 19.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 1.1 | 68.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 1.0 | 26.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.9 | 116.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.9 | 406.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.9 | 156.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.8 | 8.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.8 | 63.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.7 | 15.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.6 | 31.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.5 | 6.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.5 | 54.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.4 | 191.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.3 | 116.8 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.5 | 1037.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 13.3 | 570.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 6.9 | 124.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 6.5 | 494.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 3.6 | 157.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 3.4 | 168.2 | PID BMP PATHWAY | BMP receptor signaling |
| 3.4 | 63.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 2.8 | 187.1 | PID FGF PATHWAY | FGF signaling pathway |
| 1.8 | 55.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 1.6 | 28.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 1.2 | 76.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 1.2 | 277.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 1.2 | 84.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 1.0 | 68.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.8 | 41.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.6 | 37.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.5 | 74.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.5 | 4.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 8.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 3.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 70.6 | 2754.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 27.9 | 445.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 23.9 | 406.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 22.3 | 200.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 18.3 | 366.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 15.6 | 124.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 14.4 | 187.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 13.2 | 515.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 7.5 | 90.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 6.1 | 206.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 6.0 | 107.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 5.7 | 68.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 5.3 | 96.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 5.2 | 180.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 4.2 | 63.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 4.1 | 64.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 3.3 | 109.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 3.2 | 154.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 3.0 | 23.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 3.0 | 168.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 2.9 | 681.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 2.9 | 77.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 2.8 | 41.5 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 2.5 | 124.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 2.3 | 76.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.6 | 26.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.4 | 8.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 41.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 5.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.4 | 11.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 4.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 3.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |